|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.19 |
6.86 |
-11.68 |
2 |
4 |
0 |
58 |
291.354 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.19 |
7.13 |
-40.83 |
3 |
4 |
1 |
59 |
292.362 |
3 |
↓
|
|
|
|
Analogs
-
40861761
-
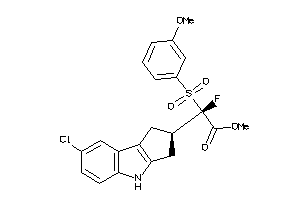
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NR1H2-1-E |
LXR-beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
56 |
0.34 |
Binding ≤ 10μM
|
|
NR1H3-1-E |
LXR-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
62 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.30 |
8.29 |
-13.16 |
1 |
6 |
0 |
85 |
451.903 |
6 |
↓
|
|
|
|
Analogs
-
40848094
-
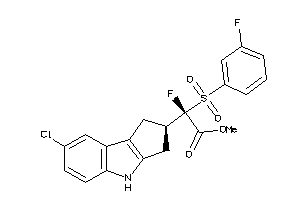
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NR1H2-1-E |
LXR-beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
360 |
0.31 |
Binding ≤ 10μM
|
|
NR1H3-1-E |
LXR-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
49 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.41 |
9.05 |
-13.56 |
1 |
5 |
0 |
76 |
439.867 |
5 |
↓
|
|
|
|
Analogs
-
40848226
-
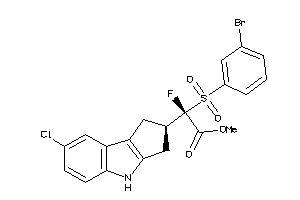
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NR1H2-1-E |
LXR-beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.36 |
Binding ≤ 10μM
|
|
NR1H3-1-E |
LXR-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
850 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.05 |
9.61 |
-12.88 |
1 |
5 |
0 |
76 |
500.773 |
5 |
↓
|
|
|
|
Analogs
-
40863096
-
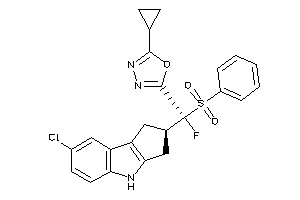
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NR1H2-1-E |
LXR-beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
92 |
0.31 |
Binding ≤ 10μM
|
|
NR1H3-1-E |
LXR-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
430 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.41 |
6.44 |
-12.74 |
1 |
6 |
0 |
89 |
471.941 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NR1H2-1-E |
LXR-beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
76 |
0.31 |
Binding ≤ 10μM
|
|
NR1H3-1-E |
LXR-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
890 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.73 |
5.62 |
-13.92 |
1 |
7 |
0 |
92 |
474.945 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NR1H2-1-E |
LXR-beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
13 |
0.35 |
Binding ≤ 10μM
|
|
NR1H3-1-E |
LXR-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
370 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.42 |
9.57 |
-13.03 |
1 |
7 |
0 |
92 |
474.945 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NR1H2-1-E |
LXR-beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
50 |
0.37 |
Binding ≤ 10μM
|
|
NR1H3-1-E |
LXR-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.27 |
8.99 |
-12.77 |
1 |
5 |
0 |
76 |
421.877 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NR1H2-1-E |
LXR-beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
700 |
0.30 |
Binding ≤ 10μM
|
|
NR1H3-1-E |
LXR-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
840 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.92 |
9.51 |
-12.97 |
1 |
5 |
0 |
76 |
456.322 |
5 |
↓
|
|
|
|
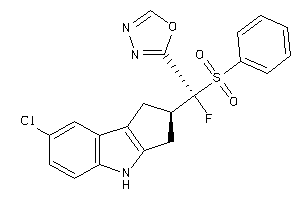
Analogs
-
40848878
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NR1H2-1-E |
LXR-beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
490 |
0.30 |
Binding ≤ 10μM
|
|
NR1H3-1-E |
LXR-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
770 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.51 |
4.09 |
-14.59 |
1 |
6 |
0 |
89 |
431.876 |
4 |
↓
|
|
|
|
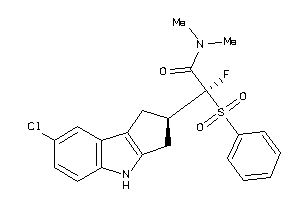
Analogs
-
40848547
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NR1H2-1-E |
LXR-beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
170 |
0.33 |
Binding ≤ 10μM
|
|
NR1H3-1-E |
LXR-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1790 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.32 |
7.99 |
-12.43 |
1 |
5 |
0 |
70 |
434.92 |
4 |
↓
|
|
|
|
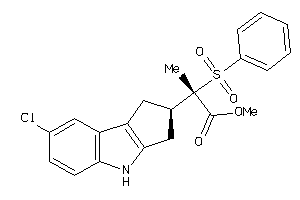
Analogs
-
40863098
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NR1H2-1-E |
LXR-beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
350 |
0.32 |
Binding ≤ 10μM
|
|
NR1H3-1-E |
LXR-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.36 |
9.76 |
-11.93 |
1 |
5 |
0 |
76 |
417.914 |
5 |
↓
|
|
|
|
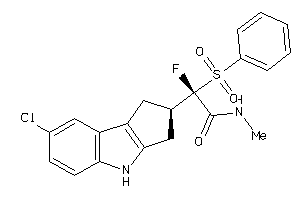
Analogs
-
40848333
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NR1H2-1-E |
LXR-beta (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
400 |
0.32 |
Binding ≤ 10μM
|
|
NR1H3-1-E |
LXR-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2700 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.51 |
6.61 |
-15.03 |
2 |
5 |
0 |
79 |
420.893 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.70 |
5.72 |
-43.49 |
1 |
5 |
-1 |
85 |
419.885 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SIRT1-1-E |
NAD-dependent Deacetylase Sirtuin 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
409 |
0.56 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.99 |
3.49 |
-11.26 |
3 |
3 |
0 |
59 |
234.686 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SIRT1-1-E |
NAD-dependent Deacetylase Sirtuin 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
409 |
0.56 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.99 |
3.54 |
-12.4 |
3 |
3 |
0 |
59 |
234.686 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SIRT1-1-E |
NAD-dependent Deacetylase Sirtuin 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2670 |
0.52 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SIRT1_HUMAN |
Q96EB6
|
NAD-dependent Deacetylase Sirtuin 1, Human |
2670 |
0.52 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.34 |
2.98 |
-12.19 |
3 |
3 |
0 |
59 |
200.241 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SIRT1-1-E |
NAD-dependent Deacetylase Sirtuin 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2670 |
0.52 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SIRT1_HUMAN |
Q96EB6
|
NAD-dependent Deacetylase Sirtuin 1, Human |
2670 |
0.52 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.34 |
3.02 |
-13.19 |
3 |
3 |
0 |
59 |
200.241 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-1-E |
Androgen Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
13 |
0.58 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.22 |
4.98 |
-6.03 |
2 |
2 |
0 |
36 |
310.102 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ANDR-1-E |
Androgen Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
13 |
0.58 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.22 |
4.98 |
-6.05 |
2 |
2 |
0 |
36 |
310.102 |
1 |
↓
|
|