|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.39 |
8.3 |
-42.87 |
1 |
2 |
1 |
22 |
210.341 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.39 |
6.09 |
-3.52 |
0 |
2 |
0 |
20 |
209.333 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.57 |
2.06 |
-61.38 |
1 |
4 |
1 |
56 |
218.298 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.57 |
-0.44 |
-15.28 |
0 |
4 |
0 |
54 |
217.29 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.29 |
4.31 |
-39.97 |
1 |
2 |
1 |
22 |
126.179 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.29 |
1.79 |
-3.98 |
0 |
2 |
0 |
20 |
125.171 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.20 |
1.73 |
-5.77 |
0 |
2 |
0 |
20 |
123.155 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.20 |
4.26 |
-41.6 |
1 |
2 |
1 |
22 |
124.163 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.44 |
5.67 |
-5.92 |
0 |
3 |
0 |
24 |
204.273 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.47 |
8 |
-7.61 |
0 |
4 |
0 |
47 |
275.348 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
26166777
-
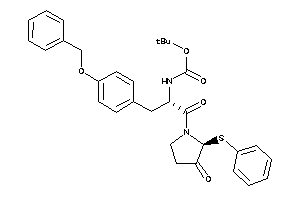
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q72874-1-V |
Human Immunodeficiency Virus Type 1 Protease (cluster #1 Of 3), Viral |
Viruses |
20 |
0.28 |
Binding ≤ 10μM
|
|
Z80295-6-O |
MT4 (Lymphocytes) (cluster #6 Of 8), Other |
Other |
100 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.06 |
14.29 |
-24.03 |
1 |
7 |
0 |
85 |
546.689 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CATB_RAT |
P00787
|
Cathepsin B, Rat |
119 |
0.26 |
Binding ≤ 1μM
|
|
CATB_HUMAN |
P07858
|
Cathepsin B, Human |
119 |
0.26 |
Binding ≤ 1μM
|
|
CATK_HUMAN |
P43235
|
Cathepsin K, Human |
3.5 |
0.32 |
Binding ≤ 1μM
|
|
CATL1_HUMAN |
P07711
|
Cathepsin L, Human |
370 |
0.24 |
Binding ≤ 1μM
|
|
CATL3_PARTE |
Q94715
|
Cathepsin L2, Parte |
370 |
0.24 |
Binding ≤ 1μM
|
|
CATS_HUMAN |
P25774
|
Cathepsin S, Human |
434 |
0.24 |
Binding ≤ 1μM
|
|
CYSP_TRYCR |
P25779
|
Cruzipain, Trycr |
93.2 |
0.27 |
Binding ≤ 1μM
|
|
LMCPB_LEIME |
P36400
|
Cysteine Proteinase B, Leime |
49.5 |
0.28 |
Binding ≤ 1μM
|
|
CATB_HUMAN |
P07858
|
Cathepsin B, Human |
119 |
0.26 |
Binding ≤ 10μM
|
|
CATB_RAT |
P00787
|
Cathepsin B, Rat |
119 |
0.26 |
Binding ≤ 10μM
|
|
CATK_HUMAN |
P43235
|
Cathepsin K, Human |
3.5 |
0.32 |
Binding ≤ 10μM
|
|
CATL1_HUMAN |
P07711
|
Cathepsin L, Human |
370 |
0.24 |
Binding ≤ 10μM
|
|
CATL3_PARTE |
Q94715
|
Cathepsin L2, Parte |
370 |
0.24 |
Binding ≤ 10μM
|
|
CATS_HUMAN |
P25774
|
Cathepsin S, Human |
434 |
0.24 |
Binding ≤ 10μM
|
|
CYSP_TRYCR |
P25779
|
Cruzipain, Trycr |
93.2 |
0.27 |
Binding ≤ 10μM
|
|
LMCPB_LEIME |
P36400
|
Cysteine Proteinase B, Leime |
49.5 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.05 |
10.33 |
-13.05 |
2 |
8 |
0 |
99 |
504.631 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CATK-1-E |
Cathepsin K (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
40 |
0.29 |
Binding ≤ 10μM
|
|
CYSP-1-E |
Cruzipain (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2500 |
0.22 |
Binding ≤ 10μM
|
|
LMCPB-1-E |
Cysteine Proteinase B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.64 |
9.34 |
-14.01 |
1 |
9 |
0 |
99 |
492.576 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.84 |
8.86 |
-68.05 |
0 |
11 |
-1 |
159 |
405.339 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.75 |
7.76 |
-149.48 |
0 |
11 |
-2 |
165 |
404.331 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.84 |
7.8 |
-61.83 |
0 |
11 |
-1 |
159 |
405.339 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.75 |
7.74 |
-139.67 |
0 |
11 |
-2 |
165 |
404.331 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.84 |
7.63 |
-69.24 |
0 |
11 |
-1 |
159 |
405.339 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.75 |
7.89 |
-146.85 |
0 |
11 |
-2 |
165 |
404.331 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.84 |
7.39 |
-67.8 |
0 |
11 |
-1 |
159 |
405.339 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.75 |
7.78 |
-149.55 |
0 |
11 |
-2 |
165 |
404.331 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.78 |
13.36 |
-26.65 |
0 |
14 |
0 |
191 |
541.469 |
12 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.92 |
12.43 |
-51.82 |
0 |
14 |
-1 |
197 |
540.461 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.78 |
12.62 |
-24.41 |
0 |
14 |
0 |
191 |
541.469 |
12 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.92 |
12.39 |
-46.64 |
0 |
14 |
-1 |
197 |
540.461 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.78 |
12.47 |
-28.42 |
0 |
14 |
0 |
191 |
541.469 |
12 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.92 |
12.55 |
-51.4 |
0 |
14 |
-1 |
197 |
540.461 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.78 |
12.23 |
-24.98 |
0 |
14 |
0 |
191 |
541.469 |
12 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.92 |
12.42 |
-51.89 |
0 |
14 |
-1 |
197 |
540.461 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.25 |
3.18 |
-67.12 |
0 |
6 |
-1 |
87 |
240.235 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.35 |
2.13 |
-152.26 |
0 |
6 |
-2 |
93 |
239.227 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.25 |
1.55 |
-58.43 |
0 |
6 |
-1 |
87 |
240.235 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.35 |
1.48 |
-139.26 |
0 |
6 |
-2 |
93 |
239.227 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.25 |
1.48 |
-65.15 |
0 |
6 |
-1 |
87 |
240.235 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.35 |
1.73 |
-146.9 |
0 |
6 |
-2 |
93 |
239.227 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.25 |
1.82 |
-64.42 |
0 |
6 |
-1 |
87 |
240.235 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.35 |
2.15 |
-152.33 |
0 |
6 |
-2 |
93 |
239.227 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.20 |
1.05 |
-8.59 |
0 |
4 |
0 |
46 |
191.234 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.20 |
3.5 |
-45.08 |
1 |
4 |
1 |
47 |
192.242 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.64 |
7.13 |
-42.79 |
1 |
2 |
1 |
22 |
222.333 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.64 |
4.8 |
-5.47 |
0 |
2 |
0 |
20 |
221.325 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.64 |
7.17 |
-40.8 |
1 |
2 |
1 |
22 |
222.333 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.64 |
4.84 |
-4.63 |
0 |
2 |
0 |
20 |
221.325 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.19 |
6.26 |
-41.85 |
1 |
2 |
1 |
22 |
196.295 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.19 |
3.95 |
-4.9 |
0 |
2 |
0 |
20 |
195.287 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.19 |
6.25 |
-40.79 |
1 |
2 |
1 |
22 |
196.295 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.19 |
3.92 |
-4.26 |
0 |
2 |
0 |
20 |
195.287 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.26 |
7.27 |
-40.28 |
1 |
2 |
1 |
22 |
196.314 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.26 |
5.16 |
-3.48 |
0 |
2 |
0 |
20 |
195.306 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.80 |
7.77 |
-39.83 |
1 |
2 |
1 |
22 |
210.341 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.80 |
5.51 |
-3.31 |
0 |
2 |
0 |
20 |
209.333 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.80 |
7.73 |
-39.98 |
1 |
2 |
1 |
22 |
210.341 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.80 |
5.6 |
-3.27 |
0 |
2 |
0 |
20 |
209.333 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.03 |
0.38 |
-38.85 |
1 |
2 |
1 |
21 |
126.179 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.03 |
0.41 |
-39.17 |
1 |
2 |
1 |
21 |
126.179 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.33 |
6.73 |
-5.77 |
0 |
2 |
0 |
20 |
247.191 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.35 |
7.65 |
-5.48 |
0 |
3 |
0 |
24 |
244.338 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.35 |
8.29 |
-32.73 |
1 |
3 |
1 |
25 |
245.346 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.85 |
6.97 |
-5.92 |
0 |
3 |
0 |
24 |
230.311 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.20 |
7.67 |
-15.85 |
1 |
3 |
0 |
25 |
233.335 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.20 |
7.57 |
-5.38 |
0 |
3 |
0 |
24 |
232.327 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.29 |
5.23 |
-6.9 |
0 |
4 |
0 |
33 |
246.31 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.84 |
2.29 |
-5.64 |
0 |
2 |
0 |
20 |
181.157 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.84 |
4.85 |
-47.08 |
1 |
2 |
1 |
22 |
182.165 |
3 |
↓
|
|
|
|
Analogs
-
39372639
-
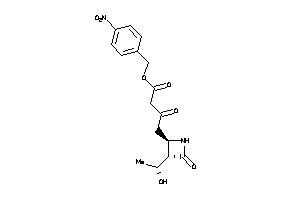
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.32 |
3.97 |
-19.18 |
1 |
9 |
0 |
130 |
348.311 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.46 |
3.73 |
-45.06 |
1 |
9 |
-1 |
136 |
347.303 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.12 |
6.42 |
-40.21 |
1 |
2 |
1 |
22 |
168.26 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.12 |
3.98 |
-3.64 |
0 |
2 |
0 |
20 |
167.252 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.12 |
6.42 |
-40.25 |
1 |
2 |
1 |
22 |
168.26 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.12 |
4.2 |
-3.68 |
0 |
2 |
0 |
20 |
167.252 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.65 |
6.67 |
-39.5 |
1 |
2 |
1 |
22 |
182.287 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.65 |
4.8 |
-3.58 |
0 |
2 |
0 |
20 |
181.279 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.65 |
6.66 |
-39.34 |
1 |
2 |
1 |
22 |
182.287 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.65 |
4.71 |
-3.48 |
0 |
2 |
0 |
20 |
181.279 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.80 |
7.1 |
-39.78 |
1 |
2 |
1 |
22 |
196.314 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.80 |
5.07 |
-3.38 |
0 |
2 |
0 |
20 |
195.306 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.80 |
7.16 |
-39.75 |
1 |
2 |
1 |
22 |
196.314 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.80 |
5.08 |
-3.43 |
0 |
2 |
0 |
20 |
195.306 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.80 |
7.12 |
-39.46 |
1 |
2 |
1 |
22 |
196.314 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.80 |
5.05 |
-3.33 |
0 |
2 |
0 |
20 |
195.306 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.80 |
6.89 |
-39.45 |
1 |
2 |
1 |
22 |
196.314 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.80 |
4.82 |
-3.33 |
0 |
2 |
0 |
20 |
195.306 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.61 |
2.54 |
-4.7 |
0 |
2 |
0 |
20 |
151.209 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.61 |
4.97 |
-37.35 |
1 |
2 |
1 |
22 |
152.217 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.07 |
4.98 |
-44.35 |
1 |
2 |
1 |
22 |
138.19 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.07 |
2.49 |
-5.09 |
0 |
2 |
0 |
20 |
137.182 |
2 |
↓
|
|