|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
36215710
-
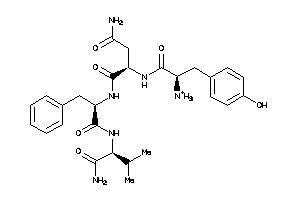
-
36215714
-
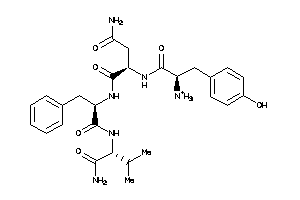
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2760 |
0.20 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
43 |
0.26 |
Binding ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
591 |
0.22 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.23 |
-1.93 |
-51.67 |
11 |
12 |
1 |
221 |
541.629 |
14 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.23 |
-2.01 |
-27.81 |
10 |
12 |
0 |
220 |
540.621 |
14 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
36216061
-
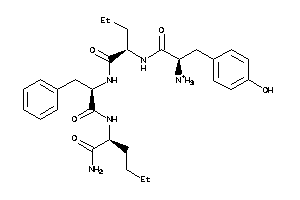
-
36216064
-
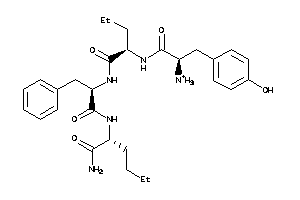
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.26 |
2.6 |
-54.86 |
9 |
10 |
1 |
178 |
540.685 |
16 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.26 |
2.28 |
-18.74 |
8 |
10 |
0 |
177 |
539.677 |
16 |
↓
|
|
|
|
|
|
|
Analogs
-
36216071
-
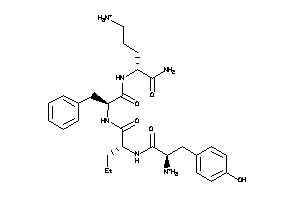
-
27757516
-
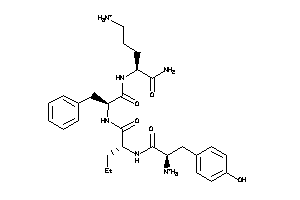
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.34 |
-0.75 |
-111.44 |
12 |
11 |
2 |
206 |
542.681 |
16 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.34 |
-1.06 |
-66.04 |
11 |
11 |
1 |
204 |
541.673 |
16 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
36216075
-
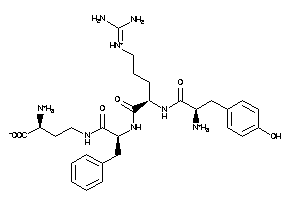
-
36216077
-
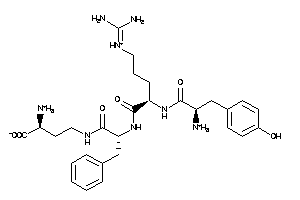
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.19 |
0.54 |
-142.35 |
15 |
14 |
2 |
269 |
586.694 |
17 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.19 |
-0.1 |
-68.92 |
13 |
14 |
0 |
266 |
584.678 |
17 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.19 |
0.23 |
-78.05 |
14 |
14 |
1 |
267 |
585.686 |
17 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.61 |
2.35 |
-123.47 |
12 |
13 |
1 |
241 |
556.644 |
16 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.61 |
2.02 |
-64.99 |
11 |
13 |
0 |
240 |
555.636 |
16 |
↓
|
|
|
|
|