Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.01 |
0.92 |
-21.34 |
3 |
9 |
0 |
125 |
352.372 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.01 |
2.14 |
-67.46 |
2 |
9 |
-1 |
127 |
351.364 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.01 |
2.16 |
-62.55 |
2 |
9 |
-1 |
127 |
351.364 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.01 |
0.94 |
-18.94 |
3 |
9 |
0 |
125 |
352.372 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.01 |
1.39 |
-53.65 |
4 |
9 |
1 |
126 |
353.38 |
4 |
↓
|
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MMP3-1-E |
Matrix Metalloproteinase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
20 |
0.45 |
Binding ≤ 10μM
|
|
MMP8-1-E |
Matrix Metalloproteinase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.49 |
Binding ≤ 10μM
|
Reactome Annotations from Targets (via Uniprot)
| Description |
Species |
|
Activation of Matrix Metalloproteinases |
|
|
Assembly of collagen fibrils and other multimeric structures |
|
|
Collagen degradation |
|
|
Degradation of the extracellular matrix |
|
|
EGFR Transactivation by Gastrin |
|
Rings
-
Glyoxaline
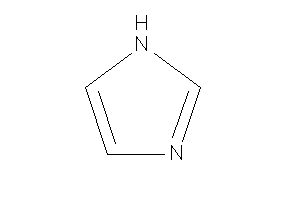
-
1,2,3,6-tetrahydropyridine
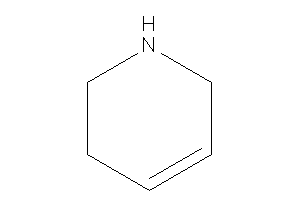
-
Benzene
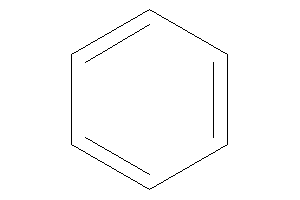
-
4,5,6,7-tetrahydro-3H-imidazo[4,…
![4,5,6,7-tetrahydro-3H-imidazo[4,5-c]pyridine](//zinc12.docking.org/img/rings/11133.gif)
-
5-besyl-3,4,6,7-tetrahydroimidaz…
![5-besyl-3,4,6,7-tetrahydroimidazo[4,5-c]pyridine](//zinc12.docking.org/img/rings/228276.gif)
No pre-computed analogs available. Try a structural similarity search.