|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FABP4-1-E |
Fatty Acid Binding Protein Adipocyte (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
670 |
0.39 |
Binding ≤ 10μM
|
|
FABP5-1-E |
Fatty Acid Binding Protein Epidermal (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3400 |
0.35 |
Binding ≤ 10μM
|
|
FABPH-1-E |
Fatty Acid Binding Protein Muscle (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9070 |
0.32 |
Binding ≤ 10μM
|
|
FABPI-1-E |
Fatty Acid Binding Protein Intestinal (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6570 |
0.33 |
Binding ≤ 10μM
|
|
FABPL-2-E |
Fatty Acid-binding Protein, Liver (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8170 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.24 |
7.54 |
-49.29 |
1 |
6 |
-1 |
96 |
336.37 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FABP4-1-E |
Fatty Acid Binding Protein Adipocyte (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.17 |
-1.17 |
-52.45 |
1 |
4 |
-1 |
69 |
293.151 |
5 |
↓
|
|
|
|
Analogs
-
4325133
-
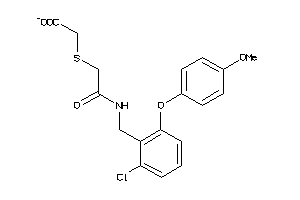
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FABP4-3-E |
Fatty Acid Binding Protein Adipocyte (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3100 |
0.32 |
Binding ≤ 10μM
|
|
FABPH-1-E |
Fatty Acid Binding Protein Muscle (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1900 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.92 |
8.86 |
-54.42 |
1 |
5 |
-1 |
78 |
364.83 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FABP4-1-E |
Fatty Acid Binding Protein Adipocyte (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4400 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.66 |
-0.29 |
-55.76 |
1 |
4 |
-1 |
69 |
276.696 |
5 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FABP4-3-E |
Fatty Acid Binding Protein Adipocyte (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3900 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.87 |
4.88 |
-15.02 |
2 |
5 |
0 |
67 |
299.252 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.32 |
3.97 |
-41.48 |
1 |
5 |
-1 |
70 |
298.244 |
5 |
↓
|
|
|
|
|
|
|
Analogs
-
36671252
-
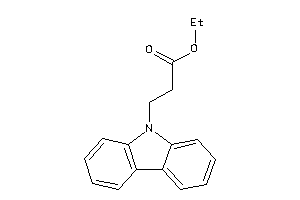
Draw
Identity
99%
90%
80%
70%
Vendors
And 31 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FABP4-3-E |
Fatty Acid Binding Protein Adipocyte (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
9400 |
0.39 |
Binding ≤ 10μM
|
|
FABPH-2-E |
Fatty Acid Binding Protein Muscle (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5700 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.05 |
0.99 |
-48.58 |
0 |
3 |
-1 |
45 |
238.266 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FABP4-3-E |
Fatty Acid Binding Protein Adipocyte (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2900 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.49 |
6 |
-14.6 |
2 |
4 |
0 |
58 |
303.671 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.95 |
5.09 |
-39.28 |
1 |
4 |
-1 |
61 |
302.663 |
4 |
↓
|
|
|
|
Analogs
-
34925820
-
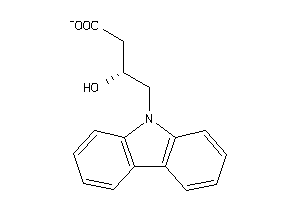
-
34925821
-
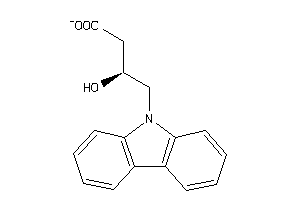
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FABP4-3-E |
Fatty Acid Binding Protein Adipocyte (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
570 |
0.46 |
Binding ≤ 10μM
|
|
FABP5-2-E |
Fatty Acid Binding Protein Epidermal (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6700 |
0.38 |
Binding ≤ 10μM
|
|
FABPH-2-E |
Fatty Acid Binding Protein Muscle (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
600 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.32 |
9.79 |
-51.27 |
0 |
3 |
-1 |
45 |
252.293 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FABP4-3-E |
Fatty Acid Binding Protein Adipocyte (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.32 |
Binding ≤ 10μM
|
|
FABP5-1-E |
Fatty Acid Binding Protein Epidermal (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
350 |
0.25 |
Binding ≤ 10μM
|
|
FABPH-1-E |
Fatty Acid Binding Protein Muscle (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3470 |
0.21 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.09 |
3.42 |
-56.54 |
0 |
5 |
-1 |
67 |
473.552 |
8 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 52 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FABP4-3-E |
Fatty Acid Binding Protein Adipocyte (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
150 |
0.45 |
Binding ≤ 10μM
|
|
TTHY-2-E |
Transthyretin (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4800 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.03 |
6.07 |
-51.55 |
1 |
4 |
-1 |
69 |
298.343 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FABP4-3-E |
Fatty Acid Binding Protein Adipocyte (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.55 |
5.16 |
-10.3 |
1 |
5 |
0 |
66 |
321.324 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.00 |
4.04 |
-39.3 |
0 |
5 |
-1 |
69 |
320.316 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 25 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FABP4-2-E |
Fatty Acid Binding Protein Adipocyte (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
930 |
0.47 |
Binding ≤ 10μM
|
|
FABP5-2-E |
Fatty Acid Binding Protein Epidermal (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
802 |
0.47 |
Binding ≤ 10μM
|
|
FABPH-3-E |
Fatty Acid Binding Protein Muscle (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2600 |
0.43 |
Binding ≤ 10μM
|
|
FABPI-2-E |
Fatty Acid Binding Protein Intestinal (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1700 |
0.45 |
Binding ≤ 10μM
|
|
TLR2-1-E |
Toll-like Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.41 |
Functional ≤ 10μM
|
|
Z80548-2-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #2 Of 5), Other |
Other |
5000 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.06 |
2.36 |
-44.91 |
0 |
2 |
-1 |
40 |
255.422 |
14 |
↓
|
|
|
|
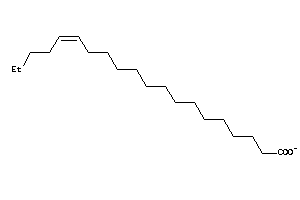
Analogs
-
33822153
-
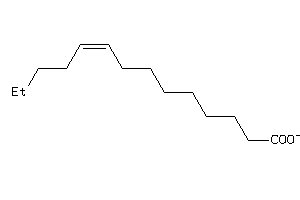
-
4529321
-
Draw
Identity
99%
90%
80%
70%
Vendors
And 18 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FAAH1-4-E |
Anandamide Amidohydrolase (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
6000 |
0.37 |
Binding ≤ 10μM
|
|
FABP4-2-E |
Fatty Acid Binding Protein Adipocyte (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
185 |
0.47 |
Binding ≤ 10μM
|
|
FABP5-2-E |
Fatty Acid Binding Protein Epidermal (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
248 |
0.46 |
Binding ≤ 10μM
|
|
FABPL-1-E |
Fatty Acid-binding Protein, Liver (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
900 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.58 |
13.34 |
-45.08 |
0 |
2 |
-1 |
40 |
281.46 |
15 |
↓
|
|