|
|
Analogs
-
12650337
-
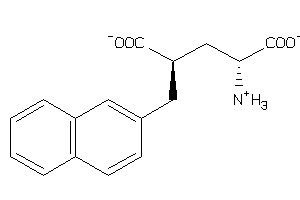
-
19969
-
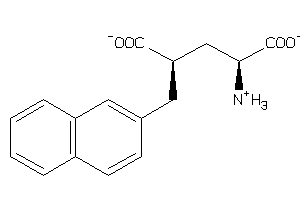
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3330 |
0.37 |
Binding ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3330 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.16 |
6.81 |
-52.05 |
3 |
5 |
-1 |
108 |
286.307 |
6 |
↓
|
|
|
|
Analogs
-
1545658
-
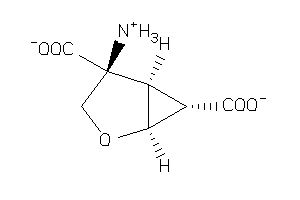
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
37 |
0.80 |
Binding ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.89 |
Binding ≤ 10μM
|
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.92 |
Functional ≤ 10μM
|
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.92 |
Functional ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.89 |
Functional ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.89 |
Functional ≤ 10μM
|
|
GRM6-1-E |
Metabotropic Glutamate Receptor 6 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.69 |
Functional ≤ 10μM
|
|
GRM6-1-E |
Metabotropic Glutamate Receptor 6 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
401 |
0.69 |
Functional ≤ 10μM
|
|
GRM8-1-E |
Metabotropic Glutamate Receptor 8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1690 |
0.62 |
Functional ≤ 10μM
|
|
GRM8-1-E |
Metabotropic Glutamate Receptor 8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1700 |
0.62 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.03 |
0.93 |
-68.12 |
3 |
6 |
-1 |
117 |
186.143 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.69 |
2.22 |
-74.96 |
3 |
5 |
-1 |
108 |
158.133 |
3 |
↓
|
|
|
|
Analogs
-
13719854
-
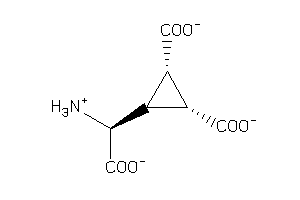
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
110 |
0.70 |
Binding ≤ 10μM
|
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.65 |
Binding ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.65 |
Binding ≤ 10μM
|
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
45 |
0.73 |
Functional ≤ 10μM
|
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.70 |
Functional ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.70 |
Functional ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
200 |
0.67 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.21 |
2.95 |
-126.68 |
3 |
7 |
-2 |
148 |
201.134 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM2-2-E |
Metabotropic Glutamate Receptor 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.38 |
Binding ≤ 10μM
|
|
GRM2-2-E |
Metabotropic Glutamate Receptor 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
110 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.62 |
13.6 |
-8.73 |
0 |
4 |
0 |
40 |
352.265 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6200 |
0.61 |
Binding ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6200 |
0.61 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.23 |
1.2 |
-80.85 |
3 |
5 |
-1 |
108 |
172.16 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6200 |
0.61 |
Binding ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6200 |
0.61 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.23 |
1.97 |
-74.44 |
3 |
5 |
-1 |
108 |
172.16 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
23 |
0.76 |
Binding ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
42 |
0.74 |
Binding ≤ 10μM
|
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
34 |
0.75 |
Functional ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
81 |
0.71 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.10 |
-1.84 |
-68.96 |
3 |
5 |
-1 |
107 |
202.161 |
2 |
↓
|
|
|
|
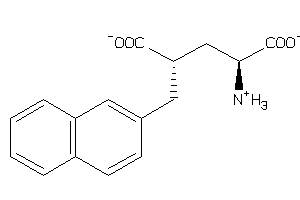
Analogs
-
13835673
-
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
48 |
0.73 |
Binding ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
66 |
0.72 |
Binding ≤ 10μM
|
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
65 |
0.72 |
Functional ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
45 |
0.73 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.95 |
-1.86 |
-73.06 |
3 |
5 |
-1 |
107 |
202.161 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.66 |
2.6 |
-75.73 |
3 |
5 |
-1 |
108 |
172.16 |
2 |
↓
|
|
|
|
Analogs
-
12650220
-
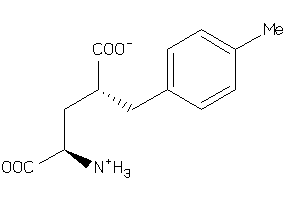
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4600 |
0.42 |
Binding ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4600 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.89 |
4.66 |
-83.69 |
3 |
5 |
-1 |
108 |
250.274 |
6 |
↓
|
|
|
|
Analogs
-
12650333
-
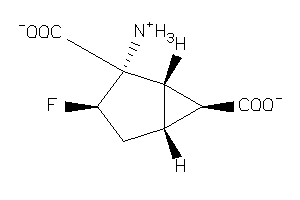
-
12650337
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8820 |
0.34 |
Binding ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8820 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.16 |
-1.62 |
-81.61 |
3 |
5 |
-1 |
107 |
286.307 |
6 |
↓
|
|
|
|
Analogs
-
12650598
-
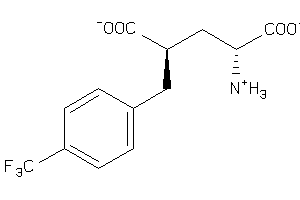
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3160 |
0.37 |
Binding ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3160 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.45 |
5.11 |
-82.8 |
3 |
5 |
-1 |
108 |
304.244 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6400 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.16 |
7.05 |
-10.76 |
2 |
3 |
0 |
49 |
250.301 |
1 |
↓
|
|
|
|
|
|
|
Analogs
-
12648269
-
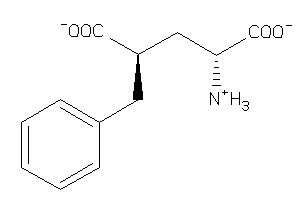
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8990 |
0.42 |
Binding ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8990 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.34 |
-1.94 |
-83.99 |
3 |
5 |
-1 |
107 |
236.247 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM1-1-E |
Metabotropic Glutamate Receptor 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8990 |
0.27 |
Binding ≤ 10μM
|
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
200 |
0.36 |
Binding ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.46 |
Binding ≤ 10μM
|
|
GRM4-2-E |
Metabotropic Glutamate Receptor 4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2650 |
0.30 |
Binding ≤ 10μM
|
|
GRM6-1-E |
Metabotropic Glutamate Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1140 |
0.32 |
Binding ≤ 10μM
|
|
GRM7-1-E |
Metabotropic Glutamate Receptor 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
110 |
0.37 |
Binding ≤ 10μM
|
|
GRM1-4-E |
Metabotropic Glutamate Receptor 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
7800 |
0.28 |
Functional ≤ 10μM
|
|
GRM2-2-E |
Metabotropic Glutamate Receptor 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
23 |
0.41 |
Functional ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
14 |
0.42 |
Functional ≤ 10μM
|
|
GRM5-2-E |
Metabotropic Glutamate Receptor 5 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
8200 |
0.27 |
Functional ≤ 10μM
|
|
GRM7-2-E |
Metabotropic Glutamate Receptor 7 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
990 |
0.32 |
Functional ≤ 10μM
|
|
GRM8-1-E |
Metabotropic Glutamate Receptor 8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
170 |
0.36 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 5), Other |
Other |
10 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.75 |
10.29 |
-80.49 |
3 |
6 |
-1 |
117 |
352.366 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 145 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA3R-6-E |
Adenosine Receptor A3 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
613 |
0.87 |
Binding ≤ 10μM
|
|
EAA3-2-E |
Excitatory Amino Acid Transporter 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
51 |
1.02 |
Binding ≤ 10μM
|
|
GRIA1-3-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1360 |
0.82 |
Binding ≤ 10μM
|
|
GRIA2-3-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
940 |
0.84 |
Binding ≤ 10μM
|
|
GRIA3-3-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
500 |
0.88 |
Binding ≤ 10μM
|
|
GRIA4-3-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
868 |
0.85 |
Binding ≤ 10μM
|
|
GRIK1-3-E |
Glutamate Receptor Ionotropic Kainate 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
701 |
0.86 |
Binding ≤ 10μM
|
|
GRIK2-2-E |
Glutamate Receptor Ionotropic Kainate 2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1106 |
0.83 |
Binding ≤ 10μM
|
|
GRIK3-2-E |
Glutamate Receptor Ionotropic Kainate 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
789 |
0.85 |
Binding ≤ 10μM
|
|
GRIK4-2-E |
Glutamate Receptor Ionotropic Kainate 4 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
700 |
0.86 |
Binding ≤ 10μM
|
|
GRIK5-2-E |
Glutamate Receptor Ionotropic Kainate 5 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
700 |
0.86 |
Binding ≤ 10μM
|
|
GRM1-3-E |
Metabotropic Glutamate Receptor 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
570 |
0.87 |
Binding ≤ 10μM
|
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7400 |
0.72 |
Binding ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.77 |
Binding ≤ 10μM
|
|
GRM4-3-E |
Metabotropic Glutamate Receptor 4 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2400 |
0.79 |
Binding ≤ 10μM
|
|
GRM5-4-E |
Metabotropic Glutamate Receptor 5 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
4100 |
0.75 |
Binding ≤ 10μM
|
|
GRM6-1-E |
Metabotropic Glutamate Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7600 |
0.72 |
Binding ≤ 10μM
|
|
GRM8-1-E |
Metabotropic Glutamate Receptor 8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3400 |
0.77 |
Binding ≤ 10μM
|
|
NMDZ1-4-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
450 |
0.89 |
Binding ≤ 10μM
|
|
GRIA1-3-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
6160 |
0.73 |
Functional ≤ 10μM
|
|
GRIA2-3-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6200 |
0.73 |
Functional ≤ 10μM
|
|
GRIA3-2-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3660 |
0.76 |
Functional ≤ 10μM
|
|
GRM1-4-E |
Metabotropic Glutamate Receptor 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
10000 |
0.70 |
Functional ≤ 10μM
|
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8500 |
0.71 |
Functional ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9000 |
0.71 |
Functional ≤ 10μM
|
|
GRM4-1-E |
Metabotropic Glutamate Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6100 |
0.73 |
Functional ≤ 10μM
|
|
GRM5-5-E |
Metabotropic Glutamate Receptor 5 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
4900 |
0.74 |
Functional ≤ 10μM
|
|
GRM6-1-E |
Metabotropic Glutamate Receptor 6 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7600 |
0.72 |
Functional ≤ 10μM
|
|
GRM8-1-E |
Metabotropic Glutamate Receptor 8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9500 |
0.70 |
Functional ≤ 10μM
|
|
NMDE1-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1800 |
0.80 |
Functional ≤ 10μM
|
|
NMDZ1-1-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1800 |
0.80 |
Functional ≤ 10μM
|
|
Z104302-3-O |
Glutamate NMDA Receptor (cluster #3 Of 7), Other |
Other |
870 |
0.85 |
Binding ≤ 10μM
|
|
Z50597-3-O |
Rattus Norvegicus (cluster #3 Of 5), Other |
Other |
170 |
0.95 |
Binding ≤ 10μM
|
|
Z50597-7-O |
Rattus Norvegicus (cluster #7 Of 12), Other |
Other |
1300 |
0.82 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.25 |
1.74 |
-73.91 |
3 |
5 |
-1 |
108 |
146.122 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
380 |
0.26 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.67 |
2.32 |
-56.7 |
0 |
4 |
-1 |
66 |
453.558 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
380 |
0.26 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.67 |
16.18 |
-56.77 |
0 |
4 |
-1 |
66 |
453.558 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 120 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MURI-2-B |
Glutamate Racemase (cluster #2 Of 2), Bacterial |
Bacteria |
5800 |
0.73 |
Binding ≤ 10μM
|
|
GRIA1-3-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1362 |
0.82 |
Binding ≤ 10μM
|
|
GRIA2-3-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
940 |
0.84 |
Binding ≤ 10μM
|
|
GRIA3-3-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
500 |
0.88 |
Binding ≤ 10μM
|
|
GRIA4-3-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
868 |
0.85 |
Binding ≤ 10μM
|
|
GRIK1-3-E |
Glutamate Receptor Ionotropic Kainate 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
701 |
0.86 |
Binding ≤ 10μM
|
|
GRIK2-2-E |
Glutamate Receptor Ionotropic Kainate 2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1106 |
0.83 |
Binding ≤ 10μM
|
|
GRIK3-2-E |
Glutamate Receptor Ionotropic Kainate 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
789 |
0.85 |
Binding ≤ 10μM
|
|
GRIK4-2-E |
Glutamate Receptor Ionotropic Kainate 4 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.90 |
Binding ≤ 10μM
|
|
GRIK5-2-E |
Glutamate Receptor Ionotropic Kainate 5 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.90 |
Binding ≤ 10μM
|
|
GRM1-3-E |
Metabotropic Glutamate Receptor 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
4900 |
0.74 |
Binding ≤ 10μM
|
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
290 |
0.92 |
Binding ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.80 |
Binding ≤ 10μM
|
|
GRM4-3-E |
Metabotropic Glutamate Receptor 4 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
9800 |
0.70 |
Binding ≤ 10μM
|
|
GRM5-4-E |
Metabotropic Glutamate Receptor 5 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
3100 |
0.77 |
Binding ≤ 10μM
|
|
GRM6-1-E |
Metabotropic Glutamate Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4900 |
0.74 |
Binding ≤ 10μM
|
|
NMDE1-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
200 |
0.94 |
Binding ≤ 10μM
|
|
NMDE2-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
200 |
0.94 |
Binding ≤ 10μM
|
|
NMDE3-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
200 |
0.94 |
Binding ≤ 10μM
|
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
200 |
0.94 |
Binding ≤ 10μM
|
|
GRIA1-3-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
214 |
0.93 |
Functional ≤ 10μM
|
|
GRIA2-3-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
214 |
0.93 |
Functional ≤ 10μM
|
|
GRIA3-2-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
214 |
0.93 |
Functional ≤ 10μM
|
|
GRIA4-2-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
214 |
0.93 |
Functional ≤ 10μM
|
|
GRM1-4-E |
Metabotropic Glutamate Receptor 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1050 |
0.84 |
Functional ≤ 10μM
|
|
GRM8-1-E |
Metabotropic Glutamate Receptor 8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8000 |
0.71 |
Functional ≤ 10μM
|
|
Z104302-3-O |
Glutamate NMDA Receptor (cluster #3 Of 7), Other |
Other |
80 |
0.99 |
Binding ≤ 10μM |
|
Z50597-7-O |
Rattus Norvegicus (cluster #7 Of 12), Other |
Other |
2500 |
0.78 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.25 |
1.72 |
-74 |
3 |
5 |
-1 |
108 |
146.122 |
4 |
↓
|
|
|
|
Analogs
-
4424547
-
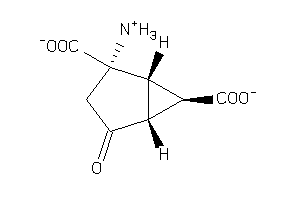
-
11679713
-
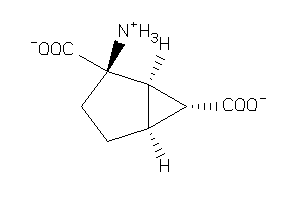
-
1547761
-
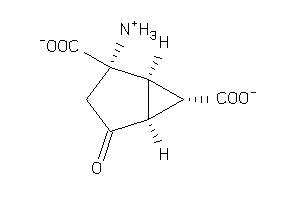
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.97 |
-2.62 |
-80.5 |
3 |
5 |
-1 |
107 |
184.171 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
819 |
0.66 |
Binding ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.87 |
Binding ≤ 10μM
|
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.90 |
Functional ≤ 10μM
|
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.90 |
Functional ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.87 |
Functional ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.87 |
Functional ≤ 10μM
|
|
GRM6-1-E |
Metabotropic Glutamate Receptor 6 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2400 |
0.61 |
Functional ≤ 10μM
|
|
GRM6-1-E |
Metabotropic Glutamate Receptor 6 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2430 |
0.60 |
Functional ≤ 10μM
|
|
GRM8-1-E |
Metabotropic Glutamate Receptor 8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7300 |
0.55 |
Functional ≤ 10μM
|
|
GRM8-1-E |
Metabotropic Glutamate Receptor 8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7340 |
0.55 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.49 |
-3.4 |
-70.4 |
3 |
5 |
-1 |
107 |
202.211 |
2 |
↓
|
|
|
|
Analogs
-
13719854
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.70 |
Functional ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.70 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.21 |
2.91 |
-132.88 |
3 |
7 |
-2 |
148 |
201.134 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GRM5-4-E |
Metabotropic Glutamate Receptor 5 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
6490 |
0.61 |
Binding ≤ 10μM
|
|
GRM6-1-E |
Metabotropic Glutamate Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6490 |
0.61 |
Binding ≤ 10μM
|
|
GRM7-1-E |
Metabotropic Glutamate Receptor 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6490 |
0.61 |
Binding ≤ 10μM
|
|
GRM8-1-E |
Metabotropic Glutamate Receptor 8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6490 |
0.61 |
Binding ≤ 10μM
|
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
450 |
0.74 |
Functional ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
410 |
0.75 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.82 |
-0.09 |
-57.41 |
4 |
6 |
-1 |
123 |
173.148 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.82 |
-1.49 |
-97.56 |
3 |
6 |
-2 |
118 |
172.14 |
2 |
↓
|
|