|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.83 |
-0.73 |
-49.96 |
4 |
7 |
1 |
92 |
422.505 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.46 |
-0.17 |
-87.78 |
3 |
7 |
0 |
98 |
421.497 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.83 |
-0.17 |
-87.78 |
3 |
7 |
0 |
94 |
421.497 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.47 |
Binding ≤ 10μM
|
|
HDA11-1-E |
Histone Deacetylase 11 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.47 |
Binding ≤ 10μM
|
|
HDAC1-2-E |
Histone Deacetylase 1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
100 |
0.47 |
Binding ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
100 |
0.47 |
Binding ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.47 |
Binding ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.47 |
Binding ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.47 |
Binding ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.47 |
Binding ≤ 10μM
|
|
HDAC7-3-E |
Histone Deacetylase 7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.47 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
100 |
0.47 |
Binding ≤ 10μM
|
|
HDAC9-3-E |
Histone Deacetylase 9 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
100 |
0.47 |
Binding ≤ 10μM
|
|
Q94F81-1-E |
Histone Deacetylase HD2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.75 |
6.51 |
-18.78 |
2 |
5 |
0 |
71 |
284.315 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.75 |
7.72 |
-58.31 |
1 |
5 |
-1 |
74 |
283.307 |
5 |
↓
|
|
|
|
Analogs
-
3872981
-
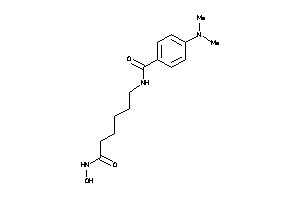
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
B1WBY8-1-E |
Histone Deacetylase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
78 |
0.45 |
Binding ≤ 10μM
|
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
46 |
0.47 |
Binding ≤ 10μM
|
|
HDA11-3-E |
Histone Deacetylase 11 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
46 |
0.47 |
Binding ≤ 10μM
|
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
78 |
0.45 |
Binding ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
46 |
0.47 |
Binding ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
78 |
0.45 |
Binding ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
78 |
0.45 |
Binding ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
46 |
0.47 |
Binding ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
750 |
0.39 |
Binding ≤ 10μM
|
|
HDAC7-3-E |
Histone Deacetylase 7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
78 |
0.45 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
46 |
0.47 |
Binding ≤ 10μM
|
|
HDAC9-3-E |
Histone Deacetylase 9 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
46 |
0.47 |
Binding ≤ 10μM
|
|
Q5RJZ2-1-E |
Histone Deacetylase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
78 |
0.45 |
Binding ≤ 10μM
|
|
Q94F81-1-E |
Histone Deacetylase HD2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.45 |
Binding ≤ 10μM
|
|
Q99P97-1-E |
Histone Deacetylase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
78 |
0.45 |
Binding ≤ 10μM
|
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1620 |
0.37 |
Functional ≤ 10μM
|
|
HDA11-1-E |
Histone Deacetylase 11 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1620 |
0.37 |
Functional ≤ 10μM
|
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1620 |
0.37 |
Functional ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1620 |
0.37 |
Functional ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1620 |
0.37 |
Functional ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1620 |
0.37 |
Functional ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1620 |
0.37 |
Functional ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1620 |
0.37 |
Functional ≤ 10μM
|
|
HDAC7-1-E |
Histone Deacetylase 7 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1620 |
0.37 |
Functional ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1620 |
0.37 |
Functional ≤ 10μM
|
|
HDAC9-1-E |
Histone Deacetylase 9 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1620 |
0.37 |
Functional ≤ 10μM
|
|
Z80532-3-O |
T-24 (Bladder Carcinoma Cells) (cluster #3 Of 6), Other |
Other |
3800 |
0.34 |
Functional ≤ 10μM
|
|
Z80682-2-O |
A549 (Lung Carcinoma Cells) (cluster #2 Of 11), Other |
Other |
800 |
0.39 |
Functional ≤ 10μM
|
|
Z80852-1-O |
A-431 (Epidermoid Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
2300 |
0.36 |
Functional ≤ 10μM
|
|
Z80877-1-O |
NCI-H1299 (Non-small Cell Lung Carcinoma) (cluster #1 Of 2), Other |
Other |
3900 |
0.34 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
210 |
0.42 |
Functional ≤ 10μM
|
|
Z81186-1-O |
LS174T (Colon Adencocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
1500 |
0.37 |
Functional ≤ 10μM
|
|
Z81331-1-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
700 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.08 |
4.23 |
-22.59 |
3 |
6 |
0 |
82 |
307.394 |
9 |
↓
|
|
Ref
Reference (pH 7)
|
2.26 |
2.24 |
-13.63 |
3 |
6 |
0 |
85 |
307.394 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
B1WBY8-1-E |
Histone Deacetylase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
515 |
0.44 |
Binding ≤ 10μM
|
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7500 |
0.36 |
Binding ≤ 10μM
|
|
HDA11-3-E |
Histone Deacetylase 11 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
7500 |
0.36 |
Binding ≤ 10μM
|
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
515 |
0.44 |
Binding ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7500 |
0.36 |
Binding ≤ 10μM
|
|
HDAC3-2-E |
Histone Deacetylase 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
515 |
0.44 |
Binding ≤ 10μM
|
|
HDAC4-2-E |
Histone Deacetylase 4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
515 |
0.44 |
Binding ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7500 |
0.36 |
Binding ≤ 10μM
|
|
HDAC6-2-E |
Histone Deacetylase 6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7500 |
0.36 |
Binding ≤ 10μM
|
|
HDAC7-3-E |
Histone Deacetylase 7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
515 |
0.44 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7500 |
0.36 |
Binding ≤ 10μM
|
|
HDAC9-3-E |
Histone Deacetylase 9 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
7500 |
0.36 |
Binding ≤ 10μM
|
|
Q5RJZ2-1-E |
Histone Deacetylase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
515 |
0.44 |
Binding ≤ 10μM
|
|
Q99P97-1-E |
Histone Deacetylase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
515 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.87 |
-1.34 |
-20.85 |
2 |
6 |
0 |
88 |
276.292 |
6 |
↓
|
|
|
|
Analogs
-
2168378
-
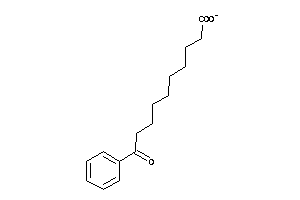
-
2378750
-
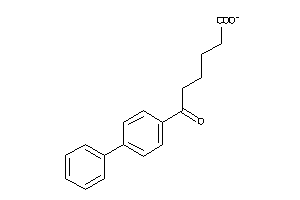
-
2378751
-
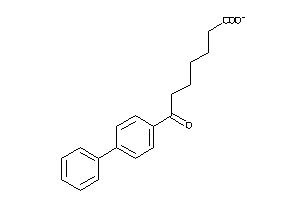
-
2579890
-
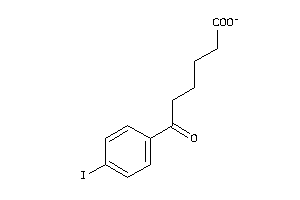
-
2579891
-
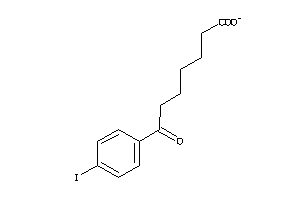
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4100 |
0.44 |
Binding ≤ 10μM
|
|
HDA11-2-E |
Histone Deacetylase 11 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4100 |
0.44 |
Binding ≤ 10μM
|
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4100 |
0.44 |
Binding ≤ 10μM
|
|
HDAC2-3-E |
Histone Deacetylase 2 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
4100 |
0.44 |
Binding ≤ 10μM
|
|
HDAC3-3-E |
Histone Deacetylase 3 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
4100 |
0.44 |
Binding ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4100 |
0.44 |
Binding ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4100 |
0.44 |
Binding ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4100 |
0.44 |
Binding ≤ 10μM
|
|
HDAC7-3-E |
Histone Deacetylase 7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
4100 |
0.44 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4100 |
0.44 |
Binding ≤ 10μM
|
|
HDAC9-3-E |
Histone Deacetylase 9 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
4100 |
0.44 |
Binding ≤ 10μM
|
|
Z80928-4-O |
HCT-116 (Colon Carcinoma Cells) (cluster #4 Of 9), Other |
Other |
1720 |
0.47 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.27 |
1.66 |
-50.41 |
0 |
3 |
-1 |
57 |
233.287 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDAC1-3-E |
Histone Deacetylase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
7000 |
1.03 |
Binding ≤ 10μM |
|
HDAC2-4-E |
Histone Deacetylase 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
9000 |
1.01 |
Binding ≤ 10μM |
|
HDAC3-3-E |
Histone Deacetylase 3 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
10000 |
1.00 |
Binding ≤ 10μM |
|
HDAC6-2-E |
Histone Deacetylase 6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2000 |
1.14 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.54 |
0.01 |
-12.32 |
2 |
3 |
0 |
49 |
103.121 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.54 |
1.22 |
-52.98 |
1 |
3 |
-1 |
52 |
102.113 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.31 |
6.66 |
-26.15 |
4 |
7 |
0 |
108 |
383.448 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.31 |
7.91 |
-62.39 |
3 |
7 |
-1 |
110 |
382.44 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
23 |
0.41 |
Binding ≤ 10μM |
|
HDA11-1-E |
Histone Deacetylase 11 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
24 |
0.41 |
Binding ≤ 10μM |
|
HDAC1-2-E |
Histone Deacetylase 1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
77 |
0.38 |
Binding ≤ 10μM |
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
27 |
0.41 |
Binding ≤ 10μM |
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
19 |
0.42 |
Binding ≤ 10μM |
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
16 |
0.42 |
Binding ≤ 10μM |
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
21 |
0.41 |
Binding ≤ 10μM |
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
247 |
0.36 |
Binding ≤ 10μM |
|
HDAC7-1-E |
Histone Deacetylase 7 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
104 |
0.38 |
Binding ≤ 10μM |
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
48 |
0.39 |
Binding ≤ 10μM |
|
HDAC9-1-E |
Histone Deacetylase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
24 |
0.41 |
Binding ≤ 10μM |
|
CP2CJ-1-E |
Cytochrome P450 2C19 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5780 |
0.28 |
ADME/T ≤ 10μM |
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2730 |
0.30 |
ADME/T ≤ 10μM |
|
Z100405-1-O |
MV4-11 (Myeloid Leukemia Cells) (cluster #1 Of 1), Other |
Other |
100 |
0.38 |
Functional ≤ 10μM |
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
340 |
0.35 |
Functional ≤ 10μM |
|
Z80415-1-O |
Ramos (Burkitts Lymhoma B-cells) (cluster #1 Of 1), Other |
Other |
140 |
0.37 |
Functional ≤ 10μM |
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
480 |
0.34 |
Functional ≤ 10μM |
|
Z81034-6-O |
A2780 (Ovarian Carcinoma Cells) (cluster #6 Of 10), Other |
Other |
480 |
0.34 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.49 |
9.6 |
-62.75 |
3 |
6 |
1 |
72 |
359.494 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.49 |
7.49 |
-18.21 |
2 |
6 |
0 |
70 |
358.486 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.49 |
9.9 |
-112.62 |
4 |
6 |
2 |
73 |
360.502 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Binding ≤ 10μM
|
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
84 |
0.43 |
Binding ≤ 10μM
|
|
HDA11-1-E |
Histone Deacetylase 11 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Binding ≤ 10μM
|
|
HDA11-1-E |
Histone Deacetylase 11 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
84 |
0.43 |
Binding ≤ 10μM
|
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Binding ≤ 10μM
|
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
84 |
0.43 |
Binding ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Binding ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
84 |
0.43 |
Binding ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Binding ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
84 |
0.43 |
Binding ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Binding ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
84 |
0.43 |
Binding ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Binding ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
84 |
0.43 |
Binding ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Binding ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
84 |
0.43 |
Binding ≤ 10μM
|
|
HDAC7-3-E |
Histone Deacetylase 7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Binding ≤ 10μM
|
|
HDAC7-3-E |
Histone Deacetylase 7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
84 |
0.43 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
84 |
0.43 |
Binding ≤ 10μM
|
|
HDAC9-3-E |
Histone Deacetylase 9 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Binding ≤ 10μM
|
|
HDAC9-3-E |
Histone Deacetylase 9 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
84 |
0.43 |
Binding ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
820 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.18 |
6.33 |
-16.37 |
3 |
5 |
0 |
78 |
312.369 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.18 |
7.54 |
-61.08 |
2 |
5 |
-1 |
81 |
311.361 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
16 |
0.47 |
Binding ≤ 10μM
|
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Binding ≤ 10μM
|
|
HDA11-1-E |
Histone Deacetylase 11 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
16 |
0.47 |
Binding ≤ 10μM
|
|
HDA11-1-E |
Histone Deacetylase 11 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Binding ≤ 10μM
|
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
16 |
0.47 |
Binding ≤ 10μM
|
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Binding ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
16 |
0.47 |
Binding ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Binding ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
16 |
0.47 |
Binding ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Binding ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
16 |
0.47 |
Binding ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Binding ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
16 |
0.47 |
Binding ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Binding ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
16 |
0.47 |
Binding ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Binding ≤ 10μM
|
|
HDAC7-3-E |
Histone Deacetylase 7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
16 |
0.47 |
Binding ≤ 10μM
|
|
HDAC7-3-E |
Histone Deacetylase 7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
16 |
0.47 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Binding ≤ 10μM
|
|
HDAC9-3-E |
Histone Deacetylase 9 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
16 |
0.47 |
Binding ≤ 10μM
|
|
HDAC9-3-E |
Histone Deacetylase 9 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
30 |
0.46 |
Binding ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
110 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.18 |
6.24 |
-21.01 |
3 |
5 |
0 |
78 |
312.369 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.18 |
7.45 |
-63.38 |
2 |
5 |
-1 |
81 |
311.361 |
5 |
↓
|
|
|
|
Analogs
-
1310
-
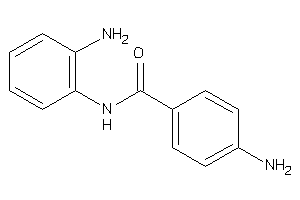
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.48 |
-2.38 |
-13.84 |
4 |
5 |
0 |
84 |
269.304 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.04 |
6.26 |
-21.31 |
4 |
5 |
0 |
84 |
351.431 |
4 |
↓
|
|
|
|
Analogs
-
33862558
-
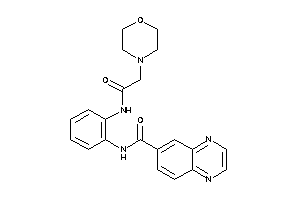
-
4860902
-
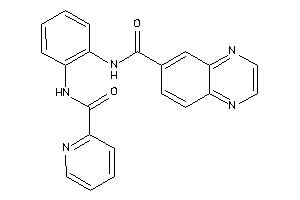
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2000 |
0.40 |
Binding ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2000 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.04 |
3.13 |
-14.61 |
3 |
5 |
0 |
81 |
264.288 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2000 |
0.44 |
Binding ≤ 10μM
|
|
HDAC2-3-E |
Histone Deacetylase 2 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
900 |
0.47 |
Binding ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1400 |
0.46 |
Binding ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
10000 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.32 |
4 |
-10.19 |
3 |
4 |
0 |
64 |
242.278 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2630 |
0.49 |
Binding ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6980 |
0.45 |
Binding ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3640 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.03 |
2.91 |
-9.74 |
3 |
4 |
0 |
68 |
213.24 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDAC1-4-E |
Histone Deacetylase 1 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
672 |
0.66 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.02 |
2.96 |
-14.92 |
2 |
3 |
0 |
49 |
193.227 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.02 |
4.2 |
-57.37 |
1 |
3 |
-1 |
52 |
192.219 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDAC1-3-E |
Histone Deacetylase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
9700 |
0.78 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.40 |
0.42 |
-17.92 |
2 |
3 |
0 |
49 |
143.167 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.40 |
1.66 |
-58.58 |
1 |
3 |
-1 |
52 |
142.159 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
715 |
0.61 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.52 |
3.82 |
-16.02 |
2 |
3 |
0 |
49 |
193.246 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4800 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.66 |
4.09 |
-10.67 |
2 |
3 |
0 |
49 |
227.263 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.94 |
8.89 |
-15.21 |
4 |
7 |
0 |
106 |
396.454 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4800 |
0.27 |
Binding ≤ 10μM
|
|
HDA11-1-E |
Histone Deacetylase 11 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4800 |
0.27 |
Binding ≤ 10μM
|
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
600 |
0.31 |
Binding ≤ 10μM
|
|
HDAC2-2-E |
Histone Deacetylase 2 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
65 |
0.36 |
Binding ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
740 |
0.31 |
Binding ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
510 |
0.31 |
Binding ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4800 |
0.27 |
Binding ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4800 |
0.27 |
Binding ≤ 10μM
|
|
HDAC7-1-E |
Histone Deacetylase 7 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4800 |
0.27 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4800 |
0.27 |
Binding ≤ 10μM
|
|
HDAC9-1-E |
Histone Deacetylase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4800 |
0.27 |
Binding ≤ 10μM
|
|
Q9XYC7-1-E |
Histone Deacetylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.30 |
Binding ≤ 10μM
|
|
TF-1-E |
Coagulation Factor III (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.26 |
Binding ≤ 10μM
|
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.30 |
Functional ≤ 10μM
|
|
HDA11-1-E |
Histone Deacetylase 11 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.30 |
Functional ≤ 10μM
|
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.30 |
Functional ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.30 |
Functional ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.30 |
Functional ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.30 |
Functional ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.30 |
Functional ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.30 |
Functional ≤ 10μM
|
|
HDAC7-1-E |
Histone Deacetylase 7 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.30 |
Functional ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.30 |
Functional ≤ 10μM
|
|
HDAC9-1-E |
Histone Deacetylase 9 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.30 |
Functional ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
730 |
0.31 |
ADME/T ≤ 10μM |
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
2000 |
0.28 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
5000 |
0.27 |
Functional ≤ 10μM
|
|
Z80269-1-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
4160 |
0.27 |
Functional ≤ 10μM
|
|
Z80475-2-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #2 Of 3), Other |
Other |
870 |
0.30 |
Functional ≤ 10μM
|
|
Z80525-1-O |
SW48 (cluster #1 Of 2), Other |
Other |
5000 |
0.27 |
Functional ≤ 10μM
|
|
Z80532-3-O |
T-24 (Bladder Carcinoma Cells) (cluster #3 Of 6), Other |
Other |
2500 |
0.28 |
Functional ≤ 10μM
|
|
Z80682-2-O |
A549 (Lung Carcinoma Cells) (cluster #2 Of 11), Other |
Other |
8000 |
0.25 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
800 |
0.30 |
Functional ≤ 10μM
|
|
Z81020-4-O |
HepG2 (Hepatoblastoma Cells) (cluster #4 Of 8), Other |
Other |
1 |
0.45 |
Functional ≤ 10μM
|
|
Z81034-3-O |
A2780 (Ovarian Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
3000 |
0.28 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
360 |
0.32 |
Functional ≤ 10μM
|
|
Z81199-1-O |
NCI-H446 (Small Cell Lung Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
1000 |
0.30 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
1800 |
0.29 |
Functional ≤ 10μM
|
|
Z81252-7-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #7 Of 11), Other |
Other |
3000 |
0.28 |
Functional ≤ 10μM
|
|
Z81331-1-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
3200 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.03 |
5.3 |
-15.7 |
4 |
7 |
0 |
106 |
376.416 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.03 |
5.76 |
-45.32 |
5 |
7 |
1 |
108 |
377.424 |
7 |
↓
|
|
|
|
Analogs
-
5178518
-
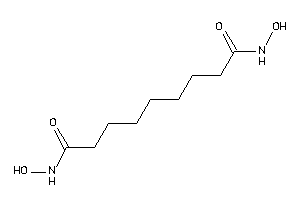
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
B1WBY8-1-E |
Histone Deacetylase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1150 |
0.59 |
Binding ≤ 10μM
|
|
HDAC1-3-E |
Histone Deacetylase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1150 |
0.59 |
Binding ≤ 10μM
|
|
HDAC2-4-E |
Histone Deacetylase 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
29 |
0.75 |
Binding ≤ 10μM
|
|
HDAC3-2-E |
Histone Deacetylase 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1150 |
0.59 |
Binding ≤ 10μM
|
|
HDAC4-2-E |
Histone Deacetylase 4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1150 |
0.59 |
Binding ≤ 10μM
|
|
HDAC5-2-E |
Histone Deacetylase 5 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9500 |
0.50 |
Binding ≤ 10μM
|
|
HDAC6-2-E |
Histone Deacetylase 6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
15 |
0.78 |
Binding ≤ 10μM
|
|
HDAC7-3-E |
Histone Deacetylase 7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1150 |
0.59 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
950 |
0.60 |
Binding ≤ 10μM
|
|
Q5RJZ2-1-E |
Histone Deacetylase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1150 |
0.59 |
Binding ≤ 10μM
|
|
Q94F81-1-E |
Histone Deacetylase HD2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
320 |
0.65 |
Binding ≤ 10μM
|
|
Q99P97-1-E |
Histone Deacetylase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1150 |
0.59 |
Binding ≤ 10μM
|
|
Z50425-6-O |
Plasmodium Falciparum (cluster #6 Of 22), Other |
Other |
2239 |
0.56 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.45 |
-0.54 |
-22.78 |
4 |
6 |
0 |
99 |
204.226 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.45 |
0.67 |
-63.15 |
3 |
6 |
-1 |
101 |
203.218 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.45 |
1.88 |
-111.59 |
2 |
6 |
-2 |
104 |
202.21 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7900 |
0.38 |
Binding ≤ 10μM
|
|
HDA11-3-E |
Histone Deacetylase 11 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
7900 |
0.38 |
Binding ≤ 10μM
|
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7900 |
0.38 |
Binding ≤ 10μM
|
|
HDAC2-4-E |
Histone Deacetylase 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
7900 |
0.38 |
Binding ≤ 10μM
|
|
HDAC3-2-E |
Histone Deacetylase 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.60 |
Binding ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7900 |
0.38 |
Binding ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7900 |
0.38 |
Binding ≤ 10μM
|
|
HDAC6-2-E |
Histone Deacetylase 6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7900 |
0.38 |
Binding ≤ 10μM
|
|
HDAC7-3-E |
Histone Deacetylase 7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
7900 |
0.38 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7900 |
0.38 |
Binding ≤ 10μM
|
|
HDAC9-3-E |
Histone Deacetylase 9 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
7900 |
0.38 |
Binding ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
6500 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.40 |
-2.76 |
-19.41 |
3 |
6 |
0 |
91 |
265.313 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.58 |
-2.2 |
-59.78 |
2 |
6 |
-1 |
97 |
264.305 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.40 |
Binding ≤ 10μM |
|
HDA11-1-E |
Histone Deacetylase 11 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.41 |
Binding ≤ 10μM |
|
HDAC1-2-E |
Histone Deacetylase 1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.40 |
Binding ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.41 |
Binding ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.40 |
Binding ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
500 |
0.32 |
Binding ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.41 |
Binding ≤ 10μM |
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
HDAC7-1-E |
Histone Deacetylase 7 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9500 |
0.25 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
400 |
0.32 |
Binding ≤ 10μM
|
|
HDAC9-1-E |
Histone Deacetylase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9000 |
0.25 |
Binding ≤ 10μM
|
|
Z80877-2-O |
NCI-H1299 (Non-small Cell Lung Carcinoma) (cluster #2 Of 2), Other |
Other |
150 |
0.34 |
Functional ≤ 10μM
|
|
Z80928-5-O |
HCT-116 (Colon Carcinoma Cells) (cluster #5 Of 9), Other |
Other |
50 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.58 |
-3.07 |
-53.1 |
5 |
6 |
1 |
89 |
380.468 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.58 |
-2.5 |
-87.1 |
4 |
6 |
0 |
92 |
379.46 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDAC1-2-E |
Histone Deacetylase 1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
995 |
0.16 |
Binding ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
980 |
0.16 |
Binding ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
980 |
0.16 |
Binding ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
450 |
0.17 |
Binding ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
450 |
0.17 |
Binding ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2900 |
0.15 |
Binding ≤ 10μM
|
|
HDAC7-2-E |
Histone Deacetylase 7 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8500 |
0.14 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
980 |
0.16 |
Binding ≤ 10μM
|
|
HDAC9-2-E |
Histone Deacetylase 9 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6300 |
0.14 |
Binding ≤ 10μM
|
|
Q94F81-1-E |
Histone Deacetylase HD2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.15 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.65 |
13.05 |
-31.39 |
4 |
10 |
0 |
143 |
721.876 |
16 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDA10-2-E |
Histone Deacetylase 10 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
270 |
0.42 |
Binding ≤ 10μM
|
|
HDA11-3-E |
Histone Deacetylase 11 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
270 |
0.42 |
Binding ≤ 10μM
|
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
270 |
0.42 |
Binding ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
270 |
0.42 |
Binding ≤ 10μM
|
|
HDAC3-2-E |
Histone Deacetylase 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
270 |
0.42 |
Binding ≤ 10μM
|
|
HDAC4-2-E |
Histone Deacetylase 4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
270 |
0.42 |
Binding ≤ 10μM
|
|
HDAC5-2-E |
Histone Deacetylase 5 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
270 |
0.42 |
Binding ≤ 10μM
|
|
HDAC6-2-E |
Histone Deacetylase 6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
270 |
0.42 |
Binding ≤ 10μM
|
|
HDAC7-3-E |
Histone Deacetylase 7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
270 |
0.42 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
270 |
0.42 |
Binding ≤ 10μM
|
|
HDAC9-3-E |
Histone Deacetylase 9 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
270 |
0.42 |
Binding ≤ 10μM
|
|
Z80418-9-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #9 Of 9), Other |
Other |
2710 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.91 |
6.79 |
-24.02 |
2 |
5 |
0 |
70 |
302.374 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.91 |
8.04 |
-67.48 |
1 |
5 |
-1 |
72 |
301.366 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDAC1-4-E |
Histone Deacetylase 1 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
6600 |
0.81 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.71 |
0.57 |
-12 |
2 |
3 |
0 |
49 |
143.167 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.71 |
1.79 |
-55.4 |
1 |
3 |
-1 |
52 |
142.159 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10 |
0.40 |
Binding ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
13 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.44 |
6.46 |
-20.49 |
4 |
6 |
0 |
93 |
375.428 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
450 |
0.68 |
Binding ≤ 10μM
|
|
HDA11-1-E |
Histone Deacetylase 11 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
450 |
0.68 |
Binding ≤ 10μM
|
|
HDAC1-2-E |
Histone Deacetylase 1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
450 |
0.68 |
Binding ≤ 10μM
|
|
HDAC2-2-E |
Histone Deacetylase 2 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
450 |
0.68 |
Binding ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
450 |
0.68 |
Binding ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
450 |
0.68 |
Binding ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
450 |
0.68 |
Binding ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
450 |
0.68 |
Binding ≤ 10μM
|
|
HDAC7-1-E |
Histone Deacetylase 7 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
450 |
0.68 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
450 |
0.68 |
Binding ≤ 10μM
|
|
HDAC9-1-E |
Histone Deacetylase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
450 |
0.68 |
Binding ≤ 10μM
|
|
Z102306-4-O |
Aorta (cluster #4 Of 6), Other |
Other |
5900 |
0.56 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.04 |
5.88 |
-12.29 |
1 |
1 |
0 |
20 |
226.347 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 15 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2400 |
0.49 |
Binding ≤ 10μM
|
|
HDAC2-2-E |
Histone Deacetylase 2 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3300 |
0.48 |
Binding ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2800 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.26 |
4.68 |
-10.08 |
3 |
3 |
0 |
55 |
212.252 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDA10-3-E |
Histone Deacetylase 10 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
59 |
0.46 |
Binding ≤ 10μM |
|
HDA11-1-E |
Histone Deacetylase 11 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
27 |
0.48 |
Binding ≤ 10μM |
|
HDAC1-2-E |
Histone Deacetylase 1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
63 |
0.46 |
Binding ≤ 10μM |
|
HDAC2-2-E |
Histone Deacetylase 2 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
22 |
0.49 |
Binding ≤ 10μM |
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
46 |
0.47 |
Binding ≤ 10μM
|
|
HDAC4-3-E |
Histone Deacetylase 4 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
380 |
0.41 |
Binding ≤ 10μM
|
|
HDAC5-3-E |
Histone Deacetylase 5 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
25 |
0.48 |
Binding ≤ 10μM |
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM
|
|
HDAC7-1-E |
Histone Deacetylase 7 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
75 |
0.45 |
Binding ≤ 10μM
|
|
HDAC8-2-E |
Histone Deacetylase 8 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
25 |
0.48 |
Binding ≤ 10μM
|
|
HDAC9-1-E |
Histone Deacetylase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
250 |
0.42 |
Binding ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
450 |
0.40 |
Functional ≤ 10μM |
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
600 |
0.40 |
Functional ≤ 10μM |
|
Z81034-3-O |
A2780 (Ovarian Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
670 |
0.39 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.19 |
2.64 |
-20.95 |
3 |
6 |
0 |
95 |
318.354 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.19 |
2.69 |
-59.56 |
2 |
6 |
-1 |
98 |
317.346 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.19 |
3.91 |
-108.48 |
1 |
6 |
-2 |
100 |
316.338 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDAH-1-B |
Histone Deacetylase-like Amidohydrolase (cluster #1 Of 2), Bacterial |
Bacteria |
9200 |
0.16 |
Binding ≤ 10μM
|
|
HDA10-2-E |
Histone Deacetylase 10 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.28 |
Binding ≤ 10μM
|
|
HDA11-3-E |
Histone Deacetylase 11 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.28 |
Binding ≤ 10μM
|
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
44 |
0.23 |
Binding ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.28 |
Binding ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.26 |
Binding ≤ 10μM
|
|
HDAC4-2-E |
Histone Deacetylase 4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.26 |
Binding ≤ 10μM
|
|
HDAC5-2-E |
Histone Deacetylase 5 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.28 |
Binding ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9600 |
0.16 |
Binding ≤ 10μM
|
|
HDAC7-2-E |
Histone Deacetylase 7 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.28 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
760 |
0.19 |
Binding ≤ 10μM
|
|
HDAC9-2-E |
Histone Deacetylase 9 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.28 |
Binding ≤ 10μM
|
|
NCOR2-1-E |
Nuclear Receptor Corepressor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
16 |
0.24 |
Binding ≤ 10μM
|
|
Q7YY07-1-E |
Histone Deacetylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.25 |
Binding ≤ 10μM
|
|
Q7YY76-1-E |
Histone Deacetylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.28 |
Binding ≤ 10μM
|
|
Q9XYC7-1-E |
Histone Deacetylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.28 |
Binding ≤ 10μM
|
|
TF-1-E |
Coagulation Factor III (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.21 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
33 |
0.23 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
290 |
0.20 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.41 |
12.84 |
-21.62 |
3 |
11 |
0 |
139 |
623.795 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDAC1-4-E |
Histone Deacetylase 1 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
5700 |
0.46 |
Binding ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3500 |
0.48 |
Binding ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
320 |
0.57 |
Binding ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
550 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.73 |
6.27 |
-4.48 |
0 |
3 |
0 |
43 |
252.213 |
5 |
↓
|
|
|
|
Analogs
-
12502280
-
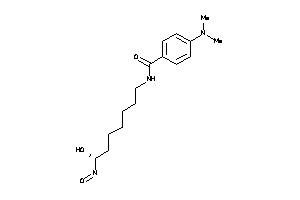
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
B1WBY8-1-E |
Histone Deacetylase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
640 |
0.41 |
Binding ≤ 10μM
|
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
244 |
0.44 |
Binding ≤ 10μM
|
|
HDA11-3-E |
Histone Deacetylase 11 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
244 |
0.44 |
Binding ≤ 10μM
|
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
640 |
0.41 |
Binding ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
244 |
0.44 |
Binding ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
640 |
0.41 |
Binding ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
640 |
0.41 |
Binding ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
244 |
0.44 |
Binding ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
244 |
0.44 |
Binding ≤ 10μM
|
|
HDAC7-3-E |
Histone Deacetylase 7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
640 |
0.41 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
244 |
0.44 |
Binding ≤ 10μM
|
|
HDAC9-3-E |
Histone Deacetylase 9 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
244 |
0.44 |
Binding ≤ 10μM
|
|
Q5RJZ2-1-E |
Histone Deacetylase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
640 |
0.41 |
Binding ≤ 10μM
|
|
Q94F81-1-E |
Histone Deacetylase HD2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.47 |
Binding ≤ 10μM
|
|
Q99P97-1-E |
Histone Deacetylase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
640 |
0.41 |
Binding ≤ 10μM
|
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
4100 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.57 |
-2.98 |
-20.1 |
3 |
6 |
0 |
81 |
293.367 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
141 |
0.50 |
Binding ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
18 |
0.57 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.77 |
5.51 |
-14.97 |
2 |
4 |
0 |
58 |
296.417 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
164 |
0.50 |
Binding ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
23 |
0.56 |
Binding ≤ 10μM
|
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
6410 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
8 |
-13.6 |
1 |
3 |
0 |
46 |
295.429 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 15 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
31 |
0.40 |
Binding ≤ 10μM |
|
HDA11-1-E |
Histone Deacetylase 11 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.45 |
Binding ≤ 10μM |
|
HDAC1-2-E |
Histone Deacetylase 1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.44 |
Binding ≤ 10μM |
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM |
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM |
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
550 |
0.34 |
Binding ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
80 |
0.38 |
Binding ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
HDAC7-1-E |
Histone Deacetylase 7 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4550 |
0.29 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
22 |
0.41 |
Binding ≤ 10μM |
|
HDAC9-1-E |
Histone Deacetylase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3200 |
0.30 |
Binding ≤ 10μM
|
|
Q9XYC7-1-E |
Histone Deacetylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
21 |
0.41 |
Functional ≤ 10μM
|
|
Z80390-3-O |
PC-3 (Prostate Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
24 |
0.41 |
Functional ≤ 10μM |
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
48 |
0.39 |
Functional ≤ 10μM |
|
Z81034-6-O |
A2780 (Ovarian Carcinoma Cells) (cluster #6 Of 10), Other |
Other |
35 |
0.40 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.19 |
6.33 |
-22.79 |
4 |
5 |
0 |
77 |
349.434 |
7 |
↓
|
|
Ref
Reference (pH 7)
|
3.37 |
5.51 |
-52.21 |
5 |
5 |
1 |
85 |
350.442 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.19 |
7.58 |
-62.39 |
3 |
5 |
-1 |
80 |
348.426 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDAC1-4-E |
Histone Deacetylase 1 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1380 |
0.48 |
Binding ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
448 |
0.52 |
Binding ≤ 10μM
|
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
4640 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.81 |
6.52 |
-7.93 |
0 |
3 |
0 |
36 |
270.375 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.51 |
4.65 |
-22.19 |
4 |
10 |
0 |
143 |
446.504 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1900 |
0.38 |
Binding ≤ 10μM
|
|
HDA11-1-E |
Histone Deacetylase 11 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1900 |
0.38 |
Binding ≤ 10μM
|
|
HDAC1-2-E |
Histone Deacetylase 1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1900 |
0.38 |
Binding ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1900 |
0.38 |
Binding ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1900 |
0.38 |
Binding ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1900 |
0.38 |
Binding ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1900 |
0.38 |
Binding ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1900 |
0.38 |
Binding ≤ 10μM
|
|
HDAC7-3-E |
Histone Deacetylase 7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1900 |
0.38 |
Binding ≤ 10μM
|
|
HDAC8-4-E |
Histone Deacetylase 8 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1900 |
0.38 |
Binding ≤ 10μM
|
|
HDAC9-3-E |
Histone Deacetylase 9 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1900 |
0.38 |
Binding ≤ 10μM
|
|
Q94F81-1-E |
Histone Deacetylase HD2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.33 |
6.15 |
-18.06 |
2 |
5 |
0 |
71 |
284.315 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.33 |
7.38 |
-57.5 |
1 |
5 |
-1 |
74 |
283.307 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDAC1-3-E |
Histone Deacetylase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
600 |
0.67 |
Binding ≤ 10μM |
|
HDAC2-4-E |
Histone Deacetylase 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
600 |
0.67 |
Binding ≤ 10μM |
|
HDAC3-2-E |
Histone Deacetylase 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.61 |
Binding ≤ 10μM |
|
HDAC6-2-E |
Histone Deacetylase 6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
500 |
0.68 |
Binding ≤ 10μM |
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4000 |
0.58 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.69 |
3.8 |
-14.65 |
2 |
3 |
0 |
49 |
179.219 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.69 |
5.02 |
-54.74 |
1 |
3 |
-1 |
52 |
178.211 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2890 |
0.24 |
Binding ≤ 10μM
|
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.38 |
Binding ≤ 10μM
|
|
ERBB2-2-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
16 |
0.34 |
Binding ≤ 10μM
|
|
FGFR2-1-E |
Fibroblast Growth Factor Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3430 |
0.24 |
Binding ≤ 10μM
|
|
FLT3-1-E |
Tyrosine-protein Kinase Receptor FLT3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.25 |
Binding ≤ 10μM
|
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
26 |
0.33 |
Binding ≤ 10μM
|
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.36 |
Binding ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
13 |
0.35 |
Binding ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.35 |
Binding ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
13 |
0.35 |
Binding ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
11 |
0.35 |
Binding ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.36 |
Binding ≤ 10μM
|
|
HDAC7-1-E |
Histone Deacetylase 7 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
373 |
0.28 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
80 |
0.31 |
Binding ≤ 10μM
|
|
HDAC9-1-E |
Histone Deacetylase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
67 |
0.31 |
Binding ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5910 |
0.23 |
Binding ≤ 10μM
|
|
LYN-1-E |
Tyrosine-protein Kinase Lyn (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
840 |
0.27 |
Binding ≤ 10μM
|
|
RET-1-E |
Tyrosine-protein Kinase Receptor RET (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3200 |
0.24 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
849 |
0.27 |
Binding ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
550 |
0.27 |
Functional ≤ 10μM
|
|
Z80322-2-O |
NCI-H358 (Lung Carcinama Cells) (cluster #2 Of 2), Other |
Other |
400 |
0.28 |
Functional ≤ 10μM
|
|
Z80475-3-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #3 Of 3), Other |
Other |
40 |
0.32 |
Functional ≤ 10μM
|
|
Z81020-4-O |
HepG2 (Hepatoblastoma Cells) (cluster #4 Of 8), Other |
Other |
130 |
0.30 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
100 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.86 |
10.33 |
-23.06 |
3 |
8 |
0 |
106 |
434.496 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDA10-3-E |
Histone Deacetylase 10 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.41 |
Binding ≤ 10μM
|
|
HDA11-1-E |
Histone Deacetylase 11 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.42 |
Binding ≤ 10μM
|
|
HDAC1-2-E |
Histone Deacetylase 1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3 |
0.41 |
Binding ≤ 10μM
|
|
HDAC2-2-E |
Histone Deacetylase 2 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.39 |
Binding ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.39 |
Binding ≤ 10μM
|
|
HDAC4-3-E |
Histone Deacetylase 4 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.40 |
Binding ≤ 10μM
|
|
HDAC5-3-E |
Histone Deacetylase 5 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.39 |
Binding ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
66 |
0.35 |
Binding ≤ 10μM
|
|
HDAC7-1-E |
Histone Deacetylase 7 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
46 |
0.35 |
Binding ≤ 10μM
|
|
HDAC8-2-E |
Histone Deacetylase 8 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
27 |
0.37 |
Binding ≤ 10μM
|
|
HDAC9-1-E |
Histone Deacetylase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
28 |
0.36 |
Binding ≤ 10μM
|
|
Z81034-1-O |
A2780 (Ovarian Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
39 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.65 |
6.15 |
-20.69 |
2 |
9 |
0 |
116 |
413.459 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.65 |
7.35 |
-58.25 |
1 |
9 |
-1 |
119 |
412.451 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 23 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
300 |
0.38 |
Binding ≤ 10μM
|
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
200 |
0.39 |
Binding ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
700 |
0.36 |
Binding ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.49 |
Binding ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7500 |
0.30 |
Binding ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
300 |
0.38 |
Binding ≤ 10μM
|
|
HDAC6-2-E |
Histone Deacetylase 6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
10 |
0.47 |
Binding ≤ 10μM
|
|
HDAC7-3-E |
Histone Deacetylase 7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2250 |
0.33 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
300 |
0.38 |
Binding ≤ 10μM
|
|
HDAC9-3-E |
Histone Deacetylase 9 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
8000 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.40 |
6.27 |
-23.43 |
2 |
6 |
0 |
88 |
326.352 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 26 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDAH-1-B |
Histone Deacetylase-like Amidohydrolase (cluster #1 Of 2), Bacterial |
Bacteria |
1300 |
0.43 |
Binding ≤ 10μM
|
|
B1WBY8-1-E |
Histone Deacetylase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
440 |
0.47 |
Binding ≤ 10μM
|
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
72 |
0.53 |
Binding ≤ 10μM
|
|
HDA11-3-E |
Histone Deacetylase 11 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
50 |
0.54 |
Binding ≤ 10μM
|
|
HDAC1-3-E |
Histone Deacetylase 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
440 |
0.47 |
Binding ≤ 10μM
|
|
HDAC2-4-E |
Histone Deacetylase 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
921 |
0.44 |
Binding ≤ 10μM |
|
HDAC3-2-E |
Histone Deacetylase 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
440 |
0.47 |
Binding ≤ 10μM
|
|
HDAC4-2-E |
Histone Deacetylase 4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
440 |
0.47 |
Binding ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
50 |
0.54 |
Binding ≤ 10μM
|
|
HDAC6-2-E |
Histone Deacetylase 6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
90 |
0.52 |
Binding ≤ 10μM
|
|
HDAC7-3-E |
Histone Deacetylase 7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
440 |
0.47 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
820 |
0.45 |
Binding ≤ 10μM
|
|
HDAC9-3-E |
Histone Deacetylase 9 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
65 |
0.53 |
Binding ≤ 10μM
|
|
NCOR2-1-E |
Nuclear Receptor Corepressor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
34 |
0.55 |
Binding ≤ 10μM
|
|
Q5RJZ2-1-E |
Histone Deacetylase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
440 |
0.47 |
Binding ≤ 10μM
|
|
Q7K6A1-1-E |
Histone Deacetylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.52 |
Binding ≤ 10μM
|
|
Q94F81-1-E |
Histone Deacetylase HD2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.54 |
Binding ≤ 10μM
|
|
Q99P97-1-E |
Histone Deacetylase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
440 |
0.47 |
Binding ≤ 10μM
|
|
Q9XYC7-1-E |
Histone Deacetylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
59 |
0.53 |
Binding ≤ 10μM
|
|
Q9ZTP8-1-E |
Histone Deacetylase HD1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
30 |
0.55 |
Binding ≤ 10μM
|
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDA11-1-E |
Histone Deacetylase 11 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDAC7-1-E |
Histone Deacetylase 7 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
HDAC9-1-E |
Histone Deacetylase 9 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Functional ≤ 10μM
|
|
Q94F81-2-E |
Histone Deacetylase HD2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.54 |
Functional ≤ 10μM
|
|
Z102015-1-O |
Plasmodium Vivax (cluster #1 Of 1), Other |
Other |
170 |
0.50 |
Functional ≤ 10μM
|
|
Z103205-2-O |
A431 (cluster #2 Of 4), Other |
Other |
4200 |
0.40 |
Functional ≤ 10μM
|
|
Z103348-2-O |
NCI-H2122 (cluster #2 Of 2), Other |
Other |
7500 |
0.38 |
Functional ≤ 10μM
|
|
Z50136-2-O |
Plasmodium Falciparum (isolate FcB1 / Columbia) (cluster #2 Of 3), Other |
Other |
820 |
0.45 |
Functional ≤ 10μM
|
|
Z50425-5-O |
Plasmodium Falciparum (cluster #5 Of 22), Other |
Other |
800 |
0.45 |
Functional ≤ 10μM
|
|
Z80054-4-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #4 Of 4), Other |
Other |
8100 |
0.38 |
Functional ≤ 10μM
|
|
Z80064-7-O |
CCRF-CEM (T-cell Leukemia) (cluster #7 Of 9), Other |
Other |
950 |
0.44 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
2120 |
0.42 |
Functional ≤ 10μM
|
|
Z80156-4-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #4 Of 12), Other |
Other |
1200 |
0.44 |
Functional ≤ 10μM
|
|
Z80164-1-O |
HT-1080 (Fibrosarcoma Cells) (cluster #1 Of 6), Other |
Other |
2400 |
0.41 |
Functional ≤ 10μM
|
|
Z80166-4-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #4 Of 12), Other |
Other |
2000 |
0.42 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
750 |
0.45 |
Functional ≤ 10μM
|
|
Z80224-6-O |
MCF7 (Breast Carcinoma Cells) (cluster #6 Of 14), Other |
Other |
8500 |
0.37 |
Functional ≤ 10μM
|
|
Z80244-1-O |
MDA-MB-468 (Breast Adenocarcinoma) (cluster #1 Of 7), Other |
Other |
5000 |
0.39 |
Functional ≤ 10μM
|
|
Z80322-1-O |
NCI-H358 (Lung Carcinama Cells) (cluster #1 Of 2), Other |
Other |
2500 |
0.41 |
Functional ≤ 10μM
|
|
Z80390-3-O |
PC-3 (Prostate Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
6500 |
0.38 |
Functional ≤ 10μM
|
|
Z80418-9-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #9 Of 9), Other |
Other |
760 |
0.45 |
Functional ≤ 10μM
|
|
Z80475-2-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #2 Of 3), Other |
Other |
2100 |
0.42 |
Functional ≤ 10μM
|
|
Z80493-3-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #3 Of 6), Other |
Other |
2500 |
0.41 |
Functional ≤ 10μM
|
|
Z80513-1-O |
SQ20B (cluster #1 Of 1), Other |
Other |
3000 |
0.41 |
Functional ≤ 10μM
|
|
Z80514-1-O |
St-4 (Stomach Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
5200 |
0.39 |
Functional ≤ 10μM
|
|
Z80682-2-O |
A549 (Lung Carcinoma Cells) (cluster #2 Of 11), Other |
Other |
868 |
0.45 |
Functional ≤ 10μM
|
|
Z80838-1-O |
EOL1 (Eosinophilic Cells) (cluster #1 Of 1), Other |
Other |
1000 |
0.44 |
Functional ≤ 10μM
|
|
Z80857-1-O |
Friend Leukemia Cell Line (cluster #1 Of 2), Other |
Other |
990 |
0.44 |
Functional ≤ 10μM
|
|
Z80874-3-O |
CEM (T-cell Leukemia) (cluster #3 Of 7), Other |
Other |
1900 |
0.42 |
Functional ≤ 10μM
|
|
Z80877-1-O |
NCI-H1299 (Non-small Cell Lung Carcinoma) (cluster #1 Of 2), Other |
Other |
7240 |
0.38 |
Functional ≤ 10μM
|
|
Z80896-1-O |
NCI-H69 (Small Cell Lung Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
2060 |
0.42 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
900 |
0.45 |
Functional ≤ 10μM
|
|
Z81020-4-O |
HepG2 (Hepatoblastoma Cells) (cluster #4 Of 8), Other |
Other |
9300 |
0.37 |
Functional ≤ 10μM
|
|
Z81024-2-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #2 Of 8), Other |
Other |
3448 |
0.40 |
Functional ≤ 10μM
|
|
Z81034-3-O |
A2780 (Ovarian Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
2600 |
0.41 |
Functional ≤ 10μM
|
|
Z81072-6-O |
Jurkat (Acute Leukemic T-cells) (cluster #6 Of 10), Other |
Other |
2300 |
0.42 |
Functional ≤ 10μM
|
|
Z81170-3-O |
LNCaP (Prostate Carcinoma) (cluster #3 Of 5), Other |
Other |
580 |
0.46 |
Functional ≤ 10μM
|
|
Z81184-5-O |
LOX IMVI (Melanoma Cells) (cluster #5 Of 5), Other |
Other |
1300 |
0.43 |
Functional ≤ 10μM
|
|
Z81245-4-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #4 Of 6), Other |
Other |
1900 |
0.42 |
Functional ≤ 10μM
|
|
Z81247-6-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #6 Of 9), Other |
Other |
460 |
0.47 |
Functional ≤ 10μM
|
|
Z81252-6-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #6 Of 11), Other |
Other |
2200 |
0.42 |
Functional ≤ 10μM
|
|
Z81280-3-O |
NCI-H226 (Non-small Cell Lung Carcinoma Cells) (cluster #3 Of 3), Other |
Other |
3480 |
0.40 |
Functional ≤ 10μM
|
|
Z81281-4-O |
NCI-H23 (Non-small Cell Lung Carcinoma Cells) (cluster #4 Of 5), Other |
Other |
383 |
0.47 |
Functional ≤ 10μM
|
|
Z81283-1-O |
NCI-H522 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
832 |
0.45 |
Functional ≤ 10μM
|
|
Z81331-1-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
600 |
0.46 |
Functional ≤ 10μM
|
|
Z81335-2-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
3900 |
0.40 |
Functional ≤ 10μM
|
|
Z80957-2-O |
HMEC (Microvascular Endothelial Cells) (cluster #2 Of 2), Other |
Other |
1300 |
0.43 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.47 |
4.41 |
-18.87 |
3 |
5 |
0 |
78 |
264.325 |
8 |
↓
|
|
|
|
Analogs
-
5298929
-
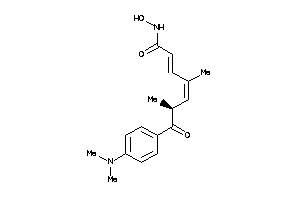
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDAH-1-B |
Histone Deacetylase-like Amidohydrolase (cluster #1 Of 2), Bacterial |
Bacteria |
700 |
0.39 |
Binding ≤ 10μM
|
|
B1WBY8-1-E |
Histone Deacetylase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
26 |
0.48 |
Binding ≤ 10μM
|
|
HDA10-3-E |
Histone Deacetylase 10 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
8 |
0.52 |
Binding ≤ 10μM
|
|
HDA11-1-E |
Histone Deacetylase 11 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Binding ≤ 10μM
|
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
26 |
0.48 |
Binding ≤ 10μM
|
|
HDAC2-2-E |
Histone Deacetylase 2 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
7600 |
0.33 |
Binding ≤ 10μM
|
|
HDAC3-2-E |
Histone Deacetylase 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
26 |
0.48 |
Binding ≤ 10μM
|
|
HDAC4-3-E |
Histone Deacetylase 4 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
26 |
0.48 |
Binding ≤ 10μM
|
|
HDAC5-3-E |
Histone Deacetylase 5 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
38 |
0.47 |
Binding ≤ 10μM
|
|
HDAC6-2-E |
Histone Deacetylase 6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
81 |
0.45 |
Binding ≤ 10μM
|
|
HDAC7-3-E |
Histone Deacetylase 7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
26 |
0.48 |
Binding ≤ 10μM
|
|
HDAC8-2-E |
Histone Deacetylase 8 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
960 |
0.38 |
Binding ≤ 10μM |
|
HDAC9-3-E |
Histone Deacetylase 9 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
800 |
0.39 |
Binding ≤ 10μM
|
|
NCOR2-2-E |
Nuclear Receptor Corepressor 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9 |
0.51 |
Binding ≤ 10μM
|
|
Q5RJZ2-1-E |
Histone Deacetylase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
26 |
0.48 |
Binding ≤ 10μM
|
|
Q94F81-1-E |
Histone Deacetylase HD2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Binding ≤ 10μM
|
|
Q99P97-1-E |
Histone Deacetylase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8600 |
0.32 |
Binding ≤ 10μM
|
|
Q9XYC7-1-E |
Histone Deacetylase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
Q9ZTP8-1-E |
Histone Deacetylase HD1B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
SF3B3-1-E |
Splicing Factor 3B Subunit 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
611 |
0.40 |
Binding ≤ 10μM
|
|
TF-1-E |
Coagulation Factor III (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.46 |
Binding ≤ 10μM
|
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.45 |
Functional ≤ 10μM
|
|
HDA11-1-E |
Histone Deacetylase 11 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.45 |
Functional ≤ 10μM
|
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.38 |
Functional ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.45 |
Functional ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.45 |
Functional ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4000 |
0.34 |
Functional ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.45 |
Functional ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.45 |
Functional ≤ 10μM
|
|
HDAC7-1-E |
Histone Deacetylase 7 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.45 |
Functional ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.45 |
Functional ≤ 10μM
|
|
HDAC9-1-E |
Histone Deacetylase 9 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.45 |
Functional ≤ 10μM
|
|
Z103359-1-O |
NCI-H661 (cluster #1 Of 1), Other |
Other |
100 |
0.45 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
10 |
0.51 |
Functional ≤ 10μM
|
|
Z80123-1-O |
DO4 (Melanoma Cells) (cluster #1 Of 1), Other |
Other |
90 |
0.45 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
60 |
0.46 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
20 |
0.49 |
Functional ≤ 10μM
|
|
Z80269-1-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
100 |
0.45 |
Functional ≤ 10μM
|
|
Z80277-1-O |
MM96L (Melanoma Cells) (cluster #1 Of 1), Other |
Other |
30 |
0.48 |
Functional ≤ 10μM
|
|
Z80475-2-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #2 Of 3), Other |
Other |
20 |
0.49 |
Functional ≤ 10μM
|
|
Z80485-1-O |
SK-MEL-28 (Melanoma Cells) (cluster #1 Of 6), Other |
Other |
280 |
0.42 |
Functional ≤ 10μM
|
|
Z80513-1-O |
SQ20B (cluster #1 Of 1), Other |
Other |
200 |
0.43 |
Functional ≤ 10μM
|
|
Z80525-1-O |
SW48 (cluster #1 Of 2), Other |
Other |
5000 |
0.34 |
Functional ≤ 10μM
|
|
Z80532-3-O |
T-24 (Bladder Carcinoma Cells) (cluster #3 Of 6), Other |
Other |
4000 |
0.34 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
80 |
0.45 |
Functional ≤ 10μM
|
|
Z80725-1-O |
C180-13S (Ovarian Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
60 |
0.46 |
Functional ≤ 10μM
|
|
Z80857-2-O |
Friend Leukemia Cell Line (cluster #2 Of 2), Other |
Other |
40 |
0.47 |
Functional ≤ 10μM
|
|
Z80877-2-O |
NCI-H1299 (Non-small Cell Lung Carcinoma) (cluster #2 Of 2), Other |
Other |
100 |
0.45 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
900 |
0.38 |
Functional ≤ 10μM
|
|
Z80957-1-O |
HMEC (Microvascular Endothelial Cells) (cluster #1 Of 2), Other |
Other |
1600 |
0.37 |
Functional ≤ 10μM
|
|
Z81070-1-O |
JAM Cell Line (cluster #1 Of 1), Other |
Other |
80 |
0.45 |
Functional ≤ 10μM
|
|
Z81199-1-O |
NCI-H446 (Small Cell Lung Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
80 |
0.45 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
90 |
0.45 |
Functional ≤ 10μM
|
|
Z80332-1-O |
NFF (Fibroblast Cells) (cluster #1 Of 1), Other |
Other |
200 |
0.43 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.68 |
6.23 |
-20.9 |
2 |
5 |
0 |
70 |
302.374 |
6 |
↓
|
|
Ref
Reference (pH 7)
|
2.68 |
6.23 |
-20.14 |
2 |
5 |
0 |
70 |
302.374 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.68 |
7.44 |
-64.15 |
1 |
5 |
-1 |
72 |
301.366 |
6 |
↓
|
|