|
|
Analogs
-
8585026
-
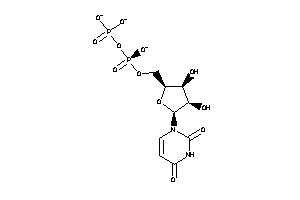
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPR17-1-E |
Uracil Nucleotide/cysteinyl Leukotriene Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1140 |
0.33 |
Binding ≤ 10μM
|
|
OGT1-1-E |
UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase 110 KDa Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1800 |
0.32 |
Binding ≤ 10μM
|
|
P2RY6-1-E |
Pyrimidinergic Receptor P2Y6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
86 |
0.40 |
Binding ≤ 10μM
|
|
P2RY6-1-E |
Pyrimidinergic Receptor P2Y6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
15 |
0.44 |
Functional ≤ 10μM
|
|
P2RY6-1-E |
Pyrimidinergic Receptor P2Y6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.38 |
Functional ≤ 10μM
|
|
P2Y14-1-E |
P2Y Purinoceptor 14 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
160 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.76 |
-4.45 |
-255.15 |
3 |
14 |
-3 |
226 |
401.137 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RY6-1-E |
Pyrimidinergic Receptor P2Y6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1720 |
0.34 |
Binding ≤ 10μM
|
|
P2RY6-1-E |
Pyrimidinergic Receptor P2Y6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1720 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RY6-1-E |
Pyrimidinergic Receptor P2Y6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
70 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.55 |
0.86 |
-264.11 |
2 |
15 |
-3 |
233 |
519.272 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.55 |
-0.3 |
-142.36 |
3 |
15 |
-2 |
230 |
520.28 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RY2-1-E |
Purinergic Receptor P2Y2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.35 |
Functional ≤ 10μM
|
|
P2RY4-1-E |
Pyrimidinergic Receptor P2Y4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
350 |
0.31 |
Functional ≤ 10μM
|
|
P2RY6-1-E |
Pyrimidinergic Receptor P2Y6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RY2-1-E |
Purinergic Receptor P2Y2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.35 |
Functional ≤ 10μM
|
|
P2RY4-1-E |
Pyrimidinergic Receptor P2Y4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
350 |
0.31 |
Functional ≤ 10μM
|
|
P2RY6-1-E |
Pyrimidinergic Receptor P2Y6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.36 |
-3.29 |
-374.05 |
3 |
16 |
-4 |
259 |
496.176 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.36 |
-4.44 |
-237.97 |
4 |
16 |
-3 |
256 |
497.184 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RY2-1-E |
Purinergic Receptor P2Y2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.35 |
Functional ≤ 10μM
|
|
P2RY4-1-E |
Pyrimidinergic Receptor P2Y4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
350 |
0.31 |
Functional ≤ 10μM
|
|
P2RY6-1-E |
Pyrimidinergic Receptor P2Y6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RY2-1-E |
Purinergic Receptor P2Y2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.35 |
Functional ≤ 10μM
|
|
P2RY4-1-E |
Pyrimidinergic Receptor P2Y4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
350 |
0.31 |
Functional ≤ 10μM
|
|
P2RY6-1-E |
Pyrimidinergic Receptor P2Y6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
-
12959016
-
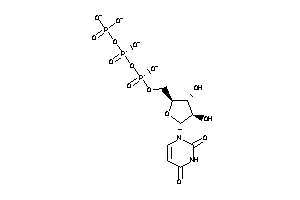
Draw
Identity
99%
90%
80%
70%
Vendors
And 27 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.57 |
-4.59 |
-370.18 |
3 |
17 |
-4 |
276 |
480.108 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.57 |
-5.75 |
-234.35 |
4 |
17 |
-3 |
273 |
481.116 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RY2-1-E |
Purinergic Receptor P2Y2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.19 |
Functional ≤ 10μM
|
|
P2RY6-1-E |
Pyrimidinergic Receptor P2Y6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1600 |
0.23 |
Functional ≤ 10μM
|
|
P2Y14-1-E |
P2Y Purinoceptor 14 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.03 |
-14.08 |
-161 |
7 |
19 |
-2 |
303 |
564.286 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RY6-1-E |
Pyrimidinergic Receptor P2Y6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
80 |
0.40 |
Functional ≤ 10μM
|
|
P2Y14-1-E |
P2Y Purinoceptor 14 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
320 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.96 |
-4.11 |
-335.1 |
2 |
13 |
-4 |
212 |
416.197 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.96 |
-5.27 |
-203.78 |
3 |
13 |
-3 |
210 |
417.205 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GPR17-1-E |
Uracil Nucleotide/cysteinyl Leukotriene Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1140 |
0.33 |
Binding ≤ 10μM
|
|
P2RY6-1-E |
Pyrimidinergic Receptor P2Y6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
15 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 25 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RY2-1-E |
Purinergic Receptor P2Y2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
130 |
0.33 |
Functional ≤ 10μM
|
|
P2RY4-1-E |
Pyrimidinergic Receptor P2Y4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
90 |
0.34 |
Functional ≤ 10μM
|
|
P2RY6-1-E |
Pyrimidinergic Receptor P2Y6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|