|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.18 |
9.62 |
-19.9 |
2 |
5 |
0 |
76 |
245.33 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
2.01 |
8.24 |
-6.23 |
2 |
5 |
0 |
73 |
245.33 |
5 |
↓
|
|
Ref
Reference (pH 7)
|
2.01 |
10.68 |
-5.25 |
2 |
5 |
0 |
73 |
245.33 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 15 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.18 |
9.68 |
-19.84 |
2 |
5 |
0 |
76 |
245.33 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
2.18 |
8.8 |
-19.26 |
2 |
5 |
0 |
76 |
245.33 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
2.01 |
8.02 |
-6.12 |
2 |
5 |
0 |
73 |
245.33 |
5 |
↓
|
|
|
|
Analogs
-
44138680
-
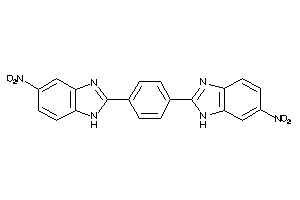
Draw
Identity
99%
90%
80%
70%
Vendors
And 18 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z102306-1-O |
Aorta (cluster #1 Of 6), Other |
Other |
4930 |
0.41 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
7.77 |
-10.72 |
1 |
5 |
0 |
75 |
239.234 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
3.48 |
7.78 |
-8.17 |
1 |
5 |
0 |
75 |
239.234 |
2 |
↓
|
|
|
|
Analogs
-
8567871
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z102306-1-O |
Aorta (cluster #1 Of 6), Other |
Other |
1810 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.82 |
4.92 |
-10.97 |
2 |
7 |
0 |
104 |
285.259 |
3 |
↓
|
|
|
|
Analogs
-
49134
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z102306-1-O |
Aorta (cluster #1 Of 6), Other |
Other |
2500 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.13 |
6.98 |
-14.18 |
1 |
7 |
0 |
93 |
299.286 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z102306-1-O |
Aorta (cluster #1 Of 6), Other |
Other |
3710 |
0.36 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.87 |
8.39 |
-13.2 |
1 |
6 |
0 |
84 |
283.287 |
4 |
↓
|
|
|
|
Analogs
-
967970
-
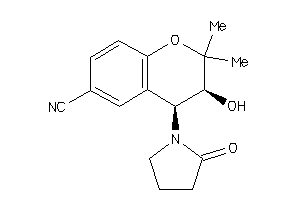
-
967971
-
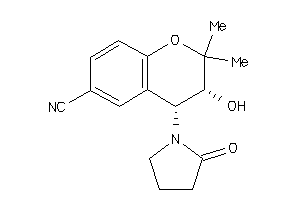
-
1224
-
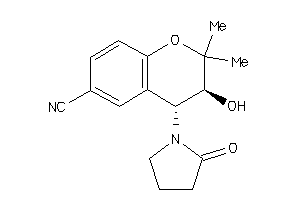
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABCC8-1-E |
Sulfonylurea Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.42 |
Binding ≤ 10μM |
|
ABCC9-1-E |
Sulfonylurea Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.42 |
Binding ≤ 10μM |
|
IRK11-1-E |
Potassium Channel, Inwardly Rectifying, Subfamily J, Member 11 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.42 |
Binding ≤ 10μM |
|
IRK8-1-E |
Potassium Channel, Inwardly Rectifying, Subfamily J, Member 8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.42 |
Binding ≤ 10μM |
|
ABCC8-1-E |
Sulfonylurea Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
570 |
0.42 |
Functional ≤ 10μM
|
|
ABCC9-1-E |
Sulfonylurea Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
570 |
0.42 |
Functional ≤ 10μM
|
|
IRK11-1-E |
Potassium Channel, Inwardly Rectifying, Subfamily J, Member 11 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
570 |
0.42 |
Functional ≤ 10μM
|
|
IRK8-1-E |
Potassium Channel, Inwardly Rectifying, Subfamily J, Member 8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
570 |
0.42 |
Functional ≤ 10μM
|
|
Z102306-1-O |
Aorta (cluster #1 Of 6), Other |
Other |
98 |
0.47 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
600 |
0.41 |
Functional ≤ 10μM |
|
Z50588-2-O |
Canis Familiaris (cluster #2 Of 7), Other |
Other |
1960 |
0.38 |
Functional ≤ 10μM |
|
Z50590-1-O |
Sus Scrofa (cluster #1 Of 1), Other |
Other |
700 |
0.41 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
420 |
0.43 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
74 |
0.48 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.41 |
5.1 |
-12.95 |
1 |
5 |
0 |
74 |
286.331 |
1 |
↓
|
|
|
|
Analogs
-
6382231
-
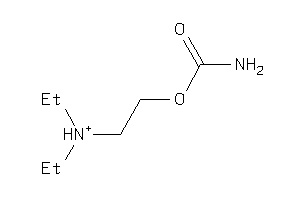
Draw
Identity
99%
90%
80%
70%
Vendors
And 31 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-8-E |
Acetylcholinesterase (cluster #8 Of 12), Eukaryotic |
Eukaryotes |
8 |
1.13 |
Binding ≤ 10μM
|
|
ACHA2-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
640 |
0.87 |
Binding ≤ 10μM
|
|
ACHB2-3-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
640 |
0.87 |
Binding ≤ 10μM
|
|
ACHB4-4-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
640 |
0.87 |
Binding ≤ 10μM
|
|
ACHD-2-E |
Acetylcholine Receptor Protein Delta Chain (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
450 |
0.89 |
Binding ≤ 10μM
|
|
ACM1-2-E |
Muscarinic Acetylcholine Receptor M1 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
739 |
0.86 |
Binding ≤ 10μM
|
|
ACM2-2-E |
Muscarinic Acetylcholine Receptor M2 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
8 |
1.13 |
Binding ≤ 10μM
|
|
ACM3-2-E |
Muscarinic Acetylcholine Receptor M3 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
9100 |
0.71 |
Binding ≤ 10μM
|
|
ACM4-2-E |
Muscarinic Acetylcholine Receptor M4 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
5500 |
0.74 |
Binding ≤ 10μM
|
|
ACM5-1-E |
Muscarinic Acetylcholine Receptor M5 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4300 |
0.75 |
Binding ≤ 10μM
|
|
ACM1-1-E |
Muscarinic Acetylcholine Receptor M1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
620 |
0.87 |
Functional ≤ 10μM
|
|
ACM2-2-E |
Muscarinic Acetylcholine Receptor M2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
700 |
0.86 |
Functional ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9900 |
0.70 |
Functional ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
230 |
0.93 |
Functional ≤ 10μM
|
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
8900 |
0.71 |
Functional ≤ 10μM
|
|
Z104281-1-O |
Neuronal Acetylcholine Receptor; Alpha3/beta2 (cluster #1 Of 2), Other |
Other |
1100 |
0.83 |
Binding ≤ 10μM
|
|
Z104286-1-O |
Neuronal Acetylcholine Receptor; Alpha2/beta2 (cluster #1 Of 2), Other |
Other |
210 |
0.93 |
Binding ≤ 10μM
|
|
Z104287-2-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #2 Of 3), Other |
Other |
4800 |
0.74 |
Binding ≤ 10μM
|
|
Z104289-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta4 (cluster #1 Of 2), Other |
Other |
1300 |
0.82 |
Binding ≤ 10μM
|
|
Z104290-3-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #3 Of 4), Other |
Other |
590 |
0.87 |
Binding ≤ 10μM
|
|
Z104303-2-O |
Muscarinic Acetylcholine Receptor (cluster #2 Of 7), Other |
Other |
9000 |
0.71 |
Binding ≤ 10μM
|
|
Z102306-2-O |
Aorta (cluster #2 Of 6), Other |
Other |
790 |
0.85 |
Functional ≤ 10μM
|
|
Z50512-2-O |
Cavia Porcellus (cluster #2 Of 7), Other |
Other |
500 |
0.88 |
Functional ≤ 10μM
|
|
Z50592-4-O |
Oryctolagus Cuniculus (cluster #4 Of 8), Other |
Other |
460 |
0.89 |
Functional ≤ 10μM
|
|
Z80024-2-O |
A9 (Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
100 |
0.98 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.14 |
1.5 |
-35.46 |
2 |
4 |
1 |
52 |
147.198 |
4 |
↓
|
|
|
|
Analogs
-
5178963
-
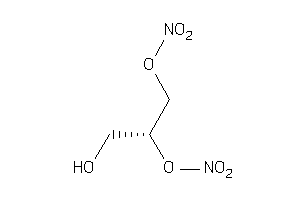
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z102306-3-O |
Aorta (cluster #3 Of 6), Other |
Other |
1700 |
0.67 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.93 |
1.06 |
-8.53 |
1 |
9 |
0 |
130 |
182.088 |
6 |
↓
|
|
|
|
Analogs
-
5178930
-
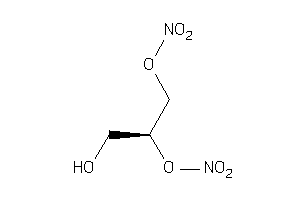
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z102306-3-O |
Aorta (cluster #3 Of 6), Other |
Other |
1700 |
0.67 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.93 |
1.06 |
-8.53 |
1 |
9 |
0 |
130 |
182.088 |
6 |
↓
|
|
|
|
Analogs
-
12353732
-
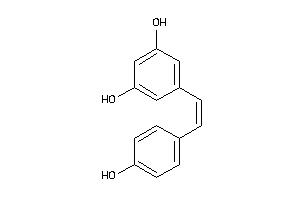
Draw
Identity
99%
90%
80%
70%
Vendors
And 55 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AHR-1-E |
Aryl Hydrocarbon Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
169 |
0.56 |
Binding ≤ 10μM
|
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
2210 |
0.47 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
950 |
0.50 |
Binding ≤ 10μM
|
|
CAH13-7-E |
Carbonic Anhydrase XIII (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
4090 |
0.44 |
Binding ≤ 10μM
|
|
CAH14-4-E |
Carbonic Anhydrase XIV (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
830 |
0.50 |
Binding ≤ 10μM
|
|
CAH15-1-E |
Carbonic Anhydrase 15 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9360 |
0.41 |
Binding ≤ 10μM
|
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
2770 |
0.46 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9090 |
0.42 |
Binding ≤ 10μM
|
|
CAH4-3-E |
Carbonic Anhydrase IV (cluster #3 Of 16), Eukaryotic |
Eukaryotes |
4470 |
0.44 |
Binding ≤ 10μM
|
|
CAH5A-8-E |
Carbonic Anhydrase VA (cluster #8 Of 10), Eukaryotic |
Eukaryotes |
4750 |
0.44 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
4640 |
0.44 |
Binding ≤ 10μM
|
|
CAH6-2-E |
Carbonic Anhydrase VI (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
8070 |
0.42 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
4350 |
0.44 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
810 |
0.50 |
Binding ≤ 10μM
|
|
ESR1-5-E |
Estrogen Receptor Alpha (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
785 |
0.50 |
Binding ≤ 10μM
|
|
NQO2-4-E |
Quinone Reductase 2 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
960 |
0.50 |
Binding ≤ 10μM
|
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
830 |
0.50 |
Binding ≤ 10μM
|
|
PGH2-1-E |
Cyclooxygenase-2 (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3490 |
0.45 |
Binding ≤ 10μM
|
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2840 |
0.46 |
Functional ≤ 10μM
|
|
Z102306-4-O |
Aorta (cluster #4 Of 6), Other |
Other |
6918 |
0.42 |
Functional ≤ 10μM
|
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
5000 |
0.44 |
Functional ≤ 10μM
|
|
Z80224-12-O |
MCF7 (Breast Carcinoma Cells) (cluster #12 Of 14), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM |
|
Z80418-1-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #1 Of 9), Other |
Other |
4000 |
0.44 |
Functional ≤ 10μM
|
|
Z80492-2-O |
SK-N-SH (Neuroblastoma Cells) (cluster #2 Of 2), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM |
|
Z80682-7-O |
A549 (Lung Carcinoma Cells) (cluster #7 Of 11), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM |
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.99 |
0.79 |
-8.46 |
3 |
3 |
0 |
61 |
228.247 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 39 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
E1BN64-1-E |
Phosphodiesterase 3B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.53 |
Binding ≤ 10μM
|
|
O77823-1-E |
Phosphodiesterase 4A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.46 |
Binding ≤ 10μM
|
|
PDE1A-1-E |
Phosphodiesterase 1A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5500 |
0.46 |
Binding ≤ 10μM
|
|
PDE3A-2-E |
Phosphodiesterase 3A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
930 |
0.53 |
Binding ≤ 10μM |
|
PDE3B-2-E |
Phosphodiesterase 3B (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
930 |
0.53 |
Binding ≤ 10μM |
|
PDE4A-3-E |
Phosphodiesterase 4A (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
6000 |
0.46 |
Binding ≤ 10μM
|
|
PDE4B-2-E |
Phosphodiesterase 4B (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.46 |
Binding ≤ 10μM
|
|
PDE4C-2-E |
Phosphodiesterase 4C (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.46 |
Binding ≤ 10μM
|
|
PDE4D-2-E |
Phosphodiesterase 4D (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.46 |
Binding ≤ 10μM
|
|
PDE5A-2-E |
Phosphodiesterase 5A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.46 |
Binding ≤ 10μM
|
|
Q864F1-1-E |
Phosphodiesterase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
740 |
0.54 |
Binding ≤ 10μM
|
|
Q9XSW7-1-E |
Phosphodiesterase 3A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
843 |
0.53 |
Binding ≤ 10μM
|
|
PGH1-1-E |
Cyclooxygenase-1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4700 |
0.47 |
Functional ≤ 10μM
|
|
Z102213-1-O |
Blood (cluster #1 Of 2), Other |
Other |
4700 |
0.47 |
Functional ≤ 10μM
|
|
Z102306-1-O |
Aorta (cluster #1 Of 6), Other |
Other |
122 |
0.60 |
Functional ≤ 10μM
|
|
Z50512-5-O |
Cavia Porcellus (cluster #5 Of 7), Other |
Other |
4700 |
0.47 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
9000 |
0.44 |
Functional ≤ 10μM
|
|
Z50588-6-O |
Canis Familiaris (cluster #6 Of 7), Other |
Other |
700 |
0.54 |
Functional ≤ 10μM
|
|
Z50589-1-O |
Felis Catus (cluster #1 Of 2), Other |
Other |
7700 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.94 |
4.19 |
-16.01 |
1 |
4 |
0 |
70 |
211.224 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.40 |
2.16 |
-43.19 |
0 |
4 |
-1 |
73 |
210.216 |
1 |
↓
|
|
|
|
Analogs
-
1802
-
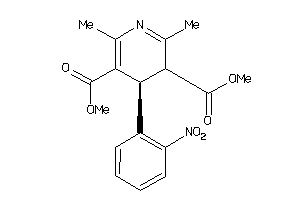
Draw
Identity
99%
90%
80%
70%
Vendors
And 24 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-1-E |
Adenosine A1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2890 |
0.31 |
Binding ≤ 10μM
|
|
AA2AR-4-E |
Adenosine A2a Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.28 |
Binding ≤ 10μM
|
|
AA3R-1-E |
Adenosine Receptor A3 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
8290 |
0.28 |
Binding ≤ 10μM
|
|
CAC1A-1-E |
Voltage-gated P/Q-type Calcium Channel Alpha-1A Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.50 |
Binding ≤ 10μM |
|
CAC1B-1-E |
Voltage-gated N-type Calcium Channel Alpha-1B Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.50 |
Binding ≤ 10μM |
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.48 |
Binding ≤ 10μM
|
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.48 |
Binding ≤ 10μM
|
|
CAC1E-1-E |
Voltage-gated R-type Calcium Channel Alpha-1E Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.50 |
Binding ≤ 10μM |
|
CAC1G-1-E |
Voltage-gated T-type Calcium Channel Alpha-1G Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.50 |
Binding ≤ 10μM |
|
CAC1H-1-E |
Voltage-gated T-type Calcium Channel Alpha-1H Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.50 |
Binding ≤ 10μM |
|
CAC1I-1-E |
Voltage-gated T-type Calcium Channel Alpha-1I Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.50 |
Binding ≤ 10μM |
|
KCNA5-1-E |
Voltage-gated Potassium Channel Subunit Kv1.5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6100 |
0.29 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
4300 |
0.30 |
Binding ≤ 10μM
|
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.48 |
Functional ≤ 10μM
|
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.48 |
Functional ≤ 10μM
|
|
TRPA1-4-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
400 |
0.36 |
Functional ≤ 10μM |
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.28 |
ADME/T ≤ 10μM
|
|
Z50592-1-O |
Oryctolagus Cuniculus (cluster #1 Of 1), Other |
Other |
1 |
0.50 |
Binding ≤ 10μM
|
|
Z102306-1-O |
Aorta (cluster #1 Of 6), Other |
Other |
20 |
0.43 |
Functional ≤ 10μM
|
|
Z102372-1-O |
Aorta (cluster #1 Of 1), Other |
Other |
9 |
0.45 |
Functional ≤ 10μM |
|
Z102380-2-O |
Ileum (cluster #2 Of 3), Other |
Other |
15 |
0.44 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7943 |
0.29 |
Functional ≤ 10μM
|
|
Z50512-1-O |
Cavia Porcellus (cluster #1 Of 7), Other |
Other |
6000 |
0.29 |
Functional ≤ 10μM
|
|
Z50590-1-O |
Sus Scrofa (cluster #1 Of 1), Other |
Other |
4 |
0.47 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
7 |
0.46 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
9 |
0.45 |
Functional ≤ 10μM
|
|
Z80051-1-O |
C6-BU-1 (cluster #1 Of 1), Other |
Other |
1000 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.32 |
8.22 |
-42.09 |
0 |
8 |
-1 |
117 |
345.331 |
5 |
↓
|
|
Ref
Reference (pH 7)
|
2.32 |
7.84 |
-40.28 |
0 |
8 |
-1 |
117 |
345.331 |
5 |
↓
|
|
|
|
Analogs
-
5071680
-
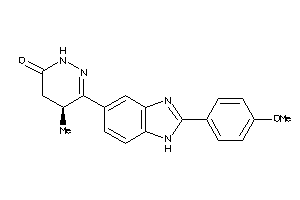
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE3A-1-E |
Phosphodiesterase 3A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
410 |
0.36 |
Binding ≤ 10μM |
|
PDE3B-1-E |
Phosphodiesterase 3B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
410 |
0.36 |
Binding ≤ 10μM |
|
Z102306-1-O |
Aorta (cluster #1 Of 6), Other |
Other |
4670 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.44 |
6.02 |
-19.35 |
2 |
6 |
0 |
79 |
334.379 |
3 |
↓
|
|
|
|
Analogs
-
4170129
-
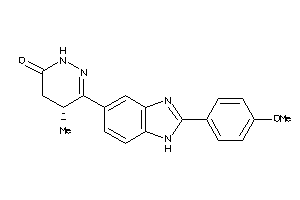
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE3A-1-E |
Phosphodiesterase 3A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
410 |
0.36 |
Binding ≤ 10μM |
|
PDE3B-1-E |
Phosphodiesterase 3B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
410 |
0.36 |
Binding ≤ 10μM |
|
Z102306-1-O |
Aorta (cluster #1 Of 6), Other |
Other |
4670 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.44 |
5.88 |
-17.98 |
2 |
6 |
0 |
79 |
334.379 |
3 |
↓
|
|
|
|
Analogs
-
4475123
-
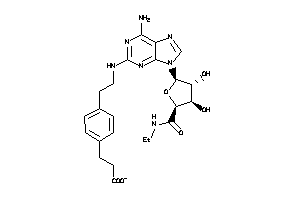
-
3872014
-
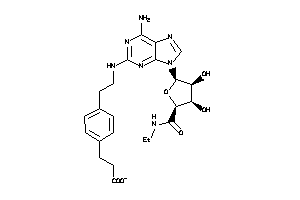
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.61 |
6.05 |
-59.85 |
6 |
13 |
-1 |
201 |
498.52 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.42 |
7.24 |
-6.72 |
0 |
1 |
0 |
17 |
194.299 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.61 |
6.43 |
-70.08 |
7 |
13 |
0 |
202 |
499.528 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDA10-1-E |
Histone Deacetylase 10 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
450 |
0.68 |
Binding ≤ 10μM
|
|
HDA11-1-E |
Histone Deacetylase 11 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
450 |
0.68 |
Binding ≤ 10μM
|
|
HDAC1-2-E |
Histone Deacetylase 1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
450 |
0.68 |
Binding ≤ 10μM
|
|
HDAC2-2-E |
Histone Deacetylase 2 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
450 |
0.68 |
Binding ≤ 10μM
|
|
HDAC3-1-E |
Histone Deacetylase 3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
450 |
0.68 |
Binding ≤ 10μM
|
|
HDAC4-1-E |
Histone Deacetylase 4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
450 |
0.68 |
Binding ≤ 10μM
|
|
HDAC5-1-E |
Histone Deacetylase 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
450 |
0.68 |
Binding ≤ 10μM
|
|
HDAC6-1-E |
Histone Deacetylase 6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
450 |
0.68 |
Binding ≤ 10μM
|
|
HDAC7-1-E |
Histone Deacetylase 7 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
450 |
0.68 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
450 |
0.68 |
Binding ≤ 10μM
|
|
HDAC9-1-E |
Histone Deacetylase 9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
450 |
0.68 |
Binding ≤ 10μM
|
|
Z102306-4-O |
Aorta (cluster #4 Of 6), Other |
Other |
5900 |
0.56 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.04 |
5.88 |
-12.29 |
1 |
1 |
0 |
20 |
226.347 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z102306-6-O |
Aorta (cluster #6 Of 6), Other |
Other |
5600 |
0.53 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.21 |
0.66 |
-12.06 |
0 |
5 |
0 |
60 |
192.174 |
2 |
↓
|
|
|
|
|
|
|
Analogs
-
34319816
-
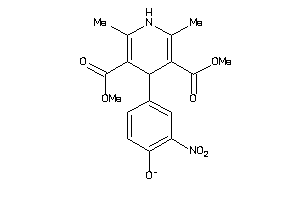
Draw
Identity
99%
90%
80%
70%
Vendors
And 20 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-1-E |
Adenosine A1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8960 |
0.27 |
Binding ≤ 10μM
|
|
AA3R-1-E |
Adenosine Receptor A3 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
8300 |
0.27 |
Binding ≤ 10μM
|
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAC1F-1-E |
Voltage-gated L-type Calcium Channel Alpha-1F Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAC1S-1-E |
Voltage-gated L-type Calcium Channel Alpha-1S Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5000 |
0.29 |
Binding ≤ 10μM
|
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.48 |
Functional ≤ 10μM
|
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.48 |
Functional ≤ 10μM
|
|
CAC1F-1-E |
Voltage-gated L-type Calcium Channel Alpha-1F Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.48 |
Functional ≤ 10μM
|
|
CAC1S-1-E |
Voltage-gated L-type Calcium Channel Alpha-1S Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.48 |
Functional ≤ 10μM
|
|
TRPA1-4-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
3800 |
0.29 |
Functional ≤ 10μM |
|
Z102306-1-O |
Aorta (cluster #1 Of 6), Other |
Other |
30 |
0.41 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7943 |
0.27 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
3 |
0.46 |
Functional ≤ 10μM
|
|
Z80387-3-O |
PC-12 (Adrenal Phaeochromacytoma Cells) (cluster #3 Of 3), Other |
Other |
10 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.19 |
9.32 |
-35.55 |
0 |
8 |
-1 |
117 |
359.358 |
6 |
↓
|
|
|
|
Analogs
-
34319816
-

-
597260
-
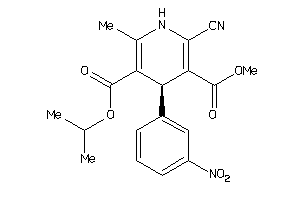
Draw
Identity
99%
90%
80%
70%
Vendors
And 30 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-1-E |
Adenosine A1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8960 |
0.27 |
Binding ≤ 10μM
|
|
AA3R-1-E |
Adenosine Receptor A3 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
8300 |
0.27 |
Binding ≤ 10μM
|
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAC1F-1-E |
Voltage-gated L-type Calcium Channel Alpha-1F Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CAC1S-1-E |
Voltage-gated L-type Calcium Channel Alpha-1S Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5000 |
0.29 |
Binding ≤ 10μM
|
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.48 |
Functional ≤ 10μM
|
|
CAC1D-1-E |
Voltage-gated L-type Calcium Channel Alpha-1D Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.48 |
Functional ≤ 10μM
|
|
CAC1F-1-E |
Voltage-gated L-type Calcium Channel Alpha-1F Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.48 |
Functional ≤ 10μM
|
|
CAC1S-1-E |
Voltage-gated L-type Calcium Channel Alpha-1S Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.48 |
Functional ≤ 10μM
|
|
TRPA1-4-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
3800 |
0.29 |
Functional ≤ 10μM |
|
Z102306-1-O |
Aorta (cluster #1 Of 6), Other |
Other |
30 |
0.41 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7943 |
0.27 |
Functional ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
3 |
0.46 |
Functional ≤ 10μM
|
|
Z80387-3-O |
PC-12 (Adrenal Phaeochromacytoma Cells) (cluster #3 Of 3), Other |
Other |
10 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.94 |
10.4 |
-17.04 |
1 |
8 |
0 |
110 |
360.366 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.82 |
8.65 |
-11.93 |
1 |
4 |
0 |
39 |
303.365 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z102306-4-O |
Aorta (cluster #4 Of 6), Other |
Other |
5790 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.85 |
7.29 |
-7.96 |
2 |
4 |
0 |
67 |
324.376 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.85 |
8.3 |
-52.44 |
1 |
4 |
-1 |
70 |
323.368 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 10 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z102306-4-O |
Aorta (cluster #4 Of 6), Other |
Other |
5790 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.85 |
7.31 |
-8.33 |
2 |
4 |
0 |
67 |
324.376 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.85 |
8.32 |
-52.3 |
1 |
4 |
-1 |
70 |
323.368 |
3 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
34663498
-
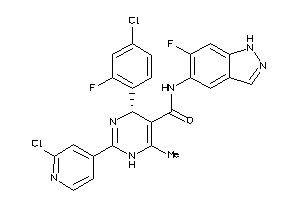
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.39 |
9.42 |
-42.33 |
4 |
7 |
1 |
97 |
514.343 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.39 |
8.3 |
-30.62 |
2 |
7 |
-1 |
97 |
512.327 |
4 |
↓
|
|
|
|
Analogs
-
34663497
-
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.39 |
8.85 |
-13.34 |
3 |
7 |
0 |
95 |
513.335 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.39 |
9.55 |
-41.97 |
4 |
7 |
1 |
97 |
514.343 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z102306-3-O |
Aorta (cluster #3 Of 6), Other |
Other |
8300 |
0.59 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.74 |
6.69 |
-43.42 |
0 |
6 |
-1 |
95 |
176.148 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.74 |
4.72 |
-7.9 |
1 |
6 |
0 |
92 |
177.156 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 73 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA2-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
180 |
0.94 |
Binding ≤ 10μM
|
|
ACHA7-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
3509 |
0.76 |
Binding ≤ 10μM
|
|
ACHB2-3-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
180 |
0.94 |
Binding ≤ 10μM
|
|
ACHB4-4-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
180 |
0.94 |
Binding ≤ 10μM
|
|
ACHD-2-E |
Acetylcholine Receptor Protein Delta Chain (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5000 |
0.74 |
Binding ≤ 10μM
|
|
ACHP-1-E |
Acetylcholine-binding Protein (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3162 |
0.77 |
Binding ≤ 10μM
|
|
ACM1-4-E |
Muscarinic Acetylcholine Receptor M1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
770 |
0.86 |
Binding ≤ 10μM
|
|
ACM3-4-E |
Muscarinic Acetylcholine Receptor M3 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
220 |
0.93 |
Binding ≤ 10μM
|
|
ACM5-1-E |
Muscarinic Acetylcholine Receptor M5 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
790 |
0.85 |
Binding ≤ 10μM
|
|
ACHA3-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8900 |
0.71 |
Functional ≤ 10μM
|
|
ACHA7-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4000 |
0.76 |
Functional ≤ 10μM
|
|
ACM1-1-E |
Muscarinic Acetylcholine Receptor M1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
1.26 |
Functional ≤ 10μM
|
|
ACM2-2-E |
Muscarinic Acetylcholine Receptor M2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
220 |
0.93 |
Functional ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
1.19 |
Functional ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
1.12 |
Functional ≤ 10μM
|
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
630 |
0.87 |
Functional ≤ 10μM
|
|
Z104281-1-O |
Neuronal Acetylcholine Receptor; Alpha3/beta2 (cluster #1 Of 2), Other |
Other |
41 |
1.03 |
Binding ≤ 10μM
|
|
Z104286-1-O |
Neuronal Acetylcholine Receptor; Alpha2/beta2 (cluster #1 Of 2), Other |
Other |
11 |
1.11 |
Binding ≤ 10μM
|
|
Z104287-2-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #2 Of 3), Other |
Other |
881 |
0.85 |
Binding ≤ 10μM
|
|
Z104289-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta4 (cluster #1 Of 2), Other |
Other |
83 |
0.99 |
Binding ≤ 10μM
|
|
Z104290-3-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #3 Of 4), Other |
Other |
8 |
1.13 |
Binding ≤ 10μM
|
|
Z104303-2-O |
Muscarinic Acetylcholine Receptor (cluster #2 Of 7), Other |
Other |
12 |
1.11 |
Binding ≤ 10μM
|
|
Z102306-2-O |
Aorta (cluster #2 Of 6), Other |
Other |
1000 |
0.84 |
Functional ≤ 10μM |
|
Z104287-1-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #1 Of 2), Other |
Other |
8700 |
0.71 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.56 |
6.42 |
-34.1 |
0 |
3 |
1 |
26 |
146.21 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.43 |
10.32 |
-28.94 |
1 |
4 |
1 |
42 |
302.357 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.24 |
8.38 |
-10.18 |
1 |
4 |
0 |
51 |
287.322 |
0 |
↓
|
|