|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 61 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
88 |
0.66 |
Binding ≤ 10μM
|
|
ACHE-1-E |
Acetylcholine Receptor Protein Epsilon Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
44 |
0.69 |
Binding ≤ 10μM
|
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
2000 |
0.53 |
Binding ≤ 10μM
|
|
ACM2-1-E |
Muscarinic Acetylcholine Receptor M2 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
220 |
0.62 |
Binding ≤ 10μM
|
|
CHLE-2-E |
Butyrylcholinesterase (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
7 |
0.76 |
Binding ≤ 10μM
|
|
CLAT-2-E |
Choline Acetylase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
HNMT-1-E |
Histamine N-methyltransferase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
110 |
0.65 |
Binding ≤ 10μM
|
|
Q9JKC1-1-E |
Butyrylcholinesterase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
92 |
0.66 |
Binding ≤ 10μM
|
|
Q9N1N9-1-E |
Butyrylcholinesterase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.76 |
Binding ≤ 10μM
|
|
ACM2-1-E |
Muscarinic Acetylcholine Receptor M2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5800 |
0.49 |
Functional ≤ 10μM
|
|
Q9WTR4-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7900 |
0.48 |
Functional ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7600 |
0.48 |
Functional ≤ 10μM
|
|
CP1A2-1-E |
Cytochrome P450 1A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1940 |
0.53 |
ADME/T ≤ 10μM
|
|
Z104298-1-O |
Glutamate NMDA Receptor; GRIN1/GRIN2A (cluster #1 Of 2), Other |
Other |
4900 |
0.50 |
Binding ≤ 10μM
|
|
Z104303-4-O |
Muscarinic Acetylcholine Receptor (cluster #4 Of 7), Other |
Other |
7300 |
0.48 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
5012 |
0.49 |
Functional ≤ 10μM
|
|
Z50594-1-O |
Mus Musculus (cluster #1 Of 9), Other |
Other |
5800 |
0.49 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.05 |
5.64 |
-23.7 |
3 |
2 |
1 |
40 |
199.277 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
2 |
0.33 |
Binding ≤ 10μM
|
|
BACE1-2-E |
Beta-secretase 1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
7500 |
0.19 |
Binding ≤ 10μM |
|
CHLE-2-E |
Butyrylcholinesterase (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
2 |
0.33 |
Binding ≤ 10μM
|
|
Q9JKC1-1-E |
Butyrylcholinesterase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
149 |
0.26 |
Binding ≤ 10μM
|
|
Q9N1N9-1-E |
Butyrylcholinesterase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.33 |
Binding ≤ 10μM
|
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
6000 |
0.20 |
Functional ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.20 |
Functional ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.20 |
Functional ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.20 |
Functional ≤ 10μM
|
|
GBRA5-3-E |
GABA Receptor Alpha-5 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.20 |
Functional ≤ 10μM
|
|
GBRA6-3-E |
GABA Receptor Alpha-6 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
6000 |
0.20 |
Functional ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.20 |
Functional ≤ 10μM
|
|
GBRB2-1-E |
GABA Receptor Beta-2 Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.20 |
Functional ≤ 10μM
|
|
GBRB3-1-E |
GABA Receptor Beta-3 Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.20 |
Functional ≤ 10μM
|
|
GBRD-1-E |
GABA Receptor Delta Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.20 |
Functional ≤ 10μM
|
|
GBRE-1-E |
GABA Receptor Epsilon Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.20 |
Functional ≤ 10μM
|
|
GBRG1-2-E |
GABA Receptor Gamma-1 Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.20 |
Functional ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6000 |
0.20 |
Functional ≤ 10μM
|
|
GBRG3-1-E |
GABA Receptor Gamma-3 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.20 |
Functional ≤ 10μM
|
|
GBRP-1-E |
GABA Receptor Pi Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.20 |
Functional ≤ 10μM
|
|
GBRT-1-E |
GABA Receptor Theta Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.20 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.47 |
17.21 |
-55.47 |
4 |
4 |
2 |
52 |
494.727 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
6289 |
0.23 |
Binding ≤ 10μM
|
|
Z104302-7-O |
Glutamate NMDA Receptor (cluster #7 Of 7), Other |
Other |
6800 |
0.23 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.00 |
13.2 |
-76.08 |
4 |
4 |
2 |
56 |
414.597 |
7 |
↓
|
|
|
|
Analogs
-
33823607
-
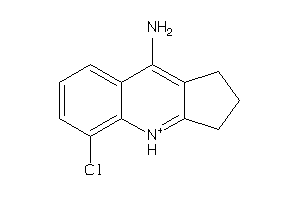
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
69 |
0.63 |
Binding ≤ 10μM |
|
CHLE-2-E |
Butyrylcholinesterase (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
45 |
0.64 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.68 |
6.27 |
-24.93 |
3 |
2 |
1 |
40 |
233.722 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.68 |
5.93 |
-10.97 |
2 |
2 |
0 |
39 |
232.714 |
0 |
↓
|
|
|
|
Analogs
-
87000
-
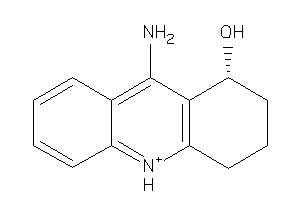
Draw
Identity
99%
90%
80%
70%
Vendors
And 17 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
4000 |
0.47 |
Binding ≤ 10μM
|
|
CHLE-2-E |
Butyrylcholinesterase (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
53 |
0.64 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.81 |
2.98 |
-24.1 |
4 |
3 |
1 |
60 |
215.276 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.81 |
2.61 |
-9.36 |
3 |
3 |
0 |
59 |
214.268 |
0 |
↓
|
|
|
|
Analogs
-
3711
-
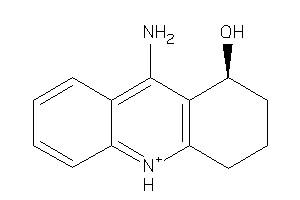
Draw
Identity
99%
90%
80%
70%
Vendors
And 17 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
4000 |
0.47 |
Binding ≤ 10μM
|
|
CHLE-2-E |
Butyrylcholinesterase (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
53 |
0.64 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.81 |
2.46 |
-24.69 |
4 |
3 |
1 |
60 |
215.276 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.81 |
2.09 |
-9.87 |
3 |
3 |
0 |
59 |
214.268 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
743 |
0.54 |
Binding ≤ 10μM |
|
CHLE-2-E |
Butyrylcholinesterase (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
1 |
0.79 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.70 |
-1.78 |
-6.11 |
2 |
2 |
0 |
38 |
232.714 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
7100 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.85 |
10.81 |
-32.73 |
0 |
5 |
1 |
38 |
296.35 |
3 |
↓
|
|
|
|
Analogs
-
4314383
-
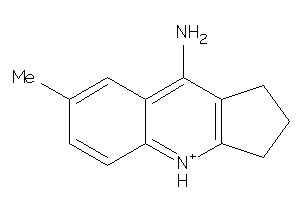
-
33823608
-
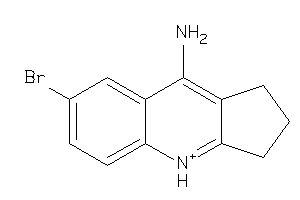
Draw
Identity
99%
90%
80%
70%
Vendors
And 39 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
56 |
0.73 |
Binding ≤ 10μM
|
|
CHLE-2-E |
Butyrylcholinesterase (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
52 |
0.73 |
Binding ≤ 10μM
|
|
Q9WTR4-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
600 |
0.62 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.53 |
5.11 |
-23.33 |
3 |
2 |
1 |
40 |
185.25 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
9100 |
0.42 |
Binding ≤ 10μM
|
|
CHLE-2-E |
Butyrylcholinesterase (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
2300 |
0.46 |
Binding ≤ 10μM
|
|
Q9WTR4-2-E |
Norepinephrine Transporter (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
800 |
0.50 |
Functional ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1100 |
0.49 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.06 |
-1.32 |
-23.77 |
3 |
2 |
1 |
40 |
227.331 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
9200 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.34 |
7.73 |
-25.39 |
2 |
3 |
1 |
35 |
271.384 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
8000 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.30 |
9.01 |
-11.72 |
1 |
3 |
0 |
36 |
316.404 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
429 |
0.59 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.15 |
4.4 |
-15.45 |
2 |
3 |
0 |
52 |
199.257 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.15 |
4.79 |
-26.55 |
3 |
3 |
1 |
53 |
200.265 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
300 |
0.35 |
Binding ≤ 10μM
|
|
CHLE-2-E |
Butyrylcholinesterase (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
2000 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.50 |
3.61 |
-73.3 |
0 |
2 |
2 |
7 |
342.486 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
9000 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.06 |
2.66 |
-84.93 |
0 |
2 |
2 |
7 |
300.405 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
4000 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.62 |
2.9 |
-73.5 |
0 |
2 |
2 |
7 |
328.459 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
1 |
0.33 |
Binding ≤ 10μM
|
|
CHLE-1-E |
Butyrylcholinesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
12 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.94 |
14.25 |
-38.6 |
4 |
6 |
1 |
95 |
529.108 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
60 |
0.34 |
Binding ≤ 10μM
|
|
ACHA7-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
7810 |
0.24 |
Binding ≤ 10μM
|
|
CHLE-2-E |
Butyrylcholinesterase (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
100 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.09 |
3.5 |
-63.39 |
0 |
2 |
2 |
7 |
398.594 |
11 |
↓
|
|
|
|
Analogs
-
42784657
-
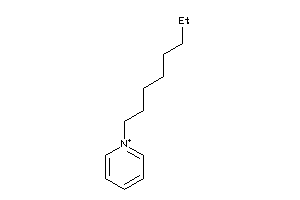
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
400 |
0.41 |
Binding ≤ 10μM
|
|
CHLE-2-E |
Butyrylcholinesterase (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
5000 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.52 |
3.9 |
-64.46 |
0 |
2 |
2 |
7 |
298.474 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
6H-Cyclohepta(b)quinoline, 7,8,9,10-tetrahydro-11-amino-; 7,8,9,10-Tetrahydro-11-amino-6H-cyclohepta(b)quinoline; BRN 0474991; LS-56080; MAS 111
6H-Cyclohepta(b)quinoline, 7,8,9…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.56 |
-1.43 |
-23.62 |
3 |
2 |
1 |
40 |
213.304 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
3 |
0.31 |
Binding ≤ 10μM
|
|
CHLE-1-E |
Butyrylcholinesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
21 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.23 |
14.3 |
-95.1 |
5 |
6 |
2 |
75 |
534.148 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
3850 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.76 |
-1.5 |
-34.3 |
1 |
4 |
1 |
42 |
266.276 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
5200 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
4.63 |
-10.2 |
2 |
4 |
0 |
57 |
258.321 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
8100 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
-1.27 |
-23.51 |
3 |
2 |
1 |
40 |
213.304 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
4 |
0.33 |
Binding ≤ 10μM
|
|
CHLE-2-E |
Butyrylcholinesterase (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
83 |
0.28 |
Binding ≤ 10μM
|
|
Q9JKC1-1-E |
Butyrylcholinesterase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
119 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.08 |
15.57 |
-57.18 |
4 |
4 |
2 |
52 |
480.7 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
4000 |
0.40 |
Binding ≤ 10μM |
|
CHLE-2-E |
Butyrylcholinesterase (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
3300 |
0.40 |
Binding ≤ 10μM
|
|
Q9JKC1-1-E |
Butyrylcholinesterase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1180 |
0.44 |
Binding ≤ 10μM |
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4600 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.65 |
-1.13 |
-24.13 |
3 |
2 |
1 |
40 |
247.321 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
2 |
0.35 |
Binding ≤ 10μM
|
|
Q9N1N9-1-E |
Butyrylcholinesterase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.69 |
13.08 |
-37.62 |
4 |
5 |
1 |
71 |
490.071 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
1 |
0.74 |
Binding ≤ 10μM
|
|
CHLE-2-E |
Butyrylcholinesterase (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
50 |
0.60 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.31 |
6.57 |
-4.12 |
2 |
2 |
0 |
39 |
267.159 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
4.31 |
6.93 |
-24.94 |
3 |
2 |
1 |
40 |
268.167 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
157 |
0.28 |
Binding ≤ 10μM
|
|
Q9JKC1-1-E |
Butyrylcholinesterase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
252 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.08 |
14.01 |
-59.96 |
4 |
4 |
2 |
52 |
452.646 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
711 |
0.27 |
Binding ≤ 10μM
|
|
Q9JKC1-1-E |
Butyrylcholinesterase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
102 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.54 |
11.97 |
-10.6 |
2 |
4 |
0 |
50 |
422.576 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
6.54 |
12.21 |
-32.59 |
3 |
4 |
1 |
51 |
423.584 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
3 |
0.31 |
Binding ≤ 10μM
|
|
CHLE-2-E |
Butyrylcholinesterase (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
3 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.74 |
16.71 |
-57.02 |
4 |
4 |
2 |
52 |
508.754 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-2-E |
Acetylcholinesterase (cluster #2 Of 12), Eukaryotic |
Eukaryotes |
31 |
0.27 |
Binding ≤ 10μM
|
|
Q9JKC1-1-E |
Butyrylcholinesterase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
155 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.96 |
17.91 |
-55.2 |
4 |
4 |
2 |
52 |
522.781 |
12 |
↓
|
|