|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 16 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A2AMW3-2-E |
GABA Receptor Epsilon Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
AA3R-3-E |
Adenosine Receptor A3 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM
|
|
FABPL-2-E |
Fatty Acid-binding Protein, Liver (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
531 |
0.44 |
Binding ≤ 10μM
|
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
5600 |
0.37 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRA5-6-E |
GABA Receptor Alpha-5 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRA6-2-E |
GABA Receptor Alpha-6 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRB2-1-E |
GABA Receptor Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
98 |
0.49 |
Binding ≤ 10μM
|
|
GBRB3-1-E |
GABA Receptor Beta-3 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRD-1-E |
GABA Receptor Delta Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRE-1-E |
GABA Receptor Epsilon Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5 |
0.58 |
Binding ≤ 10μM
|
|
GBRG1-1-E |
GABA Receptor Gamma-1 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRG3-2-E |
GABA Receptor Gamma-3 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
|
GBRP-1-E |
GABA Receptor Pi Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
12 |
0.55 |
Binding ≤ 10μM |
|
GBRR1-1-E |
GABA Receptor Rho-1 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5 |
0.58 |
Binding ≤ 10μM
|
|
GBRT-2-E |
GABA Receptor Theta Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
|
Q91ZM7-1-E |
GABA Receptor Theta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.58 |
Binding ≤ 10μM
|
|
TSPO-1-E |
Peripheral-type Benzodiazepine Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
574 |
0.44 |
Binding ≤ 10μM
|
|
GBRA1-4-E |
GABA Receptor Alpha-1 Subunit (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
2 |
0.61 |
Functional ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2 |
0.61 |
Functional ≤ 10μM
|
|
GBRG1-2-E |
GABA Receptor Gamma-1 Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.58 |
Functional ≤ 10μM
|
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 8), Other |
Other |
5 |
0.58 |
Binding ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
5 |
0.58 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.74 |
6.91 |
-8.26 |
0 |
3 |
0 |
33 |
284.746 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.74 |
7.78 |
-30.37 |
1 |
3 |
1 |
34 |
285.754 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A2AMW3-2-E |
GABA Receptor Epsilon Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.53 |
Binding ≤ 10μM
|
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
4 |
0.53 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
8 |
0.52 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
8 |
0.52 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
4 |
0.53 |
Binding ≤ 10μM
|
|
GBRA5-1-E |
GABA Receptor Alpha-5 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
4 |
0.53 |
Binding ≤ 10μM
|
|
GBRA6-2-E |
GABA Receptor Alpha-6 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
8 |
0.52 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.53 |
Binding ≤ 10μM
|
|
GBRB2-1-E |
GABA Receptor Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
4 |
0.53 |
Binding ≤ 10μM
|
|
GBRB3-1-E |
GABA Receptor Beta-3 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.53 |
Binding ≤ 10μM
|
|
GBRD-1-E |
GABA Receptor Delta Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
4 |
0.53 |
Binding ≤ 10μM
|
|
GBRE-1-E |
GABA Receptor Epsilon Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8 |
0.52 |
Binding ≤ 10μM
|
|
GBRG1-1-E |
GABA Receptor Gamma-1 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
4 |
0.53 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
4 |
0.53 |
Binding ≤ 10μM
|
|
GBRG3-2-E |
GABA Receptor Gamma-3 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
8 |
0.52 |
Binding ≤ 10μM
|
|
GBRP-1-E |
GABA Receptor Pi Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
4 |
0.53 |
Binding ≤ 10μM
|
|
GBRR1-1-E |
GABA Receptor Rho-1 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.52 |
Binding ≤ 10μM
|
|
GBRR2-1-E |
GABA Receptor Rho-2 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.52 |
Binding ≤ 10μM
|
|
GBRR3-1-E |
GABA Receptor Rho-3 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.52 |
Binding ≤ 10μM
|
|
GBRT-2-E |
GABA Receptor Theta Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
8 |
0.52 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.20 |
3.09 |
-7.34 |
2 |
4 |
0 |
67 |
367.183 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 12 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A2AMW3-2-E |
GABA Receptor Epsilon Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRA5-6-E |
GABA Receptor Alpha-5 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRA6-2-E |
GABA Receptor Alpha-6 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRB3-1-E |
GABA Receptor Beta-3 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRD-1-E |
GABA Receptor Delta Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRG1-1-E |
GABA Receptor Gamma-1 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRG3-2-E |
GABA Receptor Gamma-3 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRP-1-E |
GABA Receptor Pi Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRT-2-E |
GABA Receptor Theta Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.86 |
5.28 |
-7.85 |
1 |
3 |
0 |
41 |
270.719 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.86 |
6.15 |
-31.26 |
2 |
3 |
1 |
43 |
271.727 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
160 |
0.48 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
720 |
0.43 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
850 |
0.42 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
720 |
0.43 |
Binding ≤ 10μM
|
|
GBRA5-1-E |
GABA Receptor Alpha-5 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
561 |
0.44 |
Binding ≤ 10μM
|
|
GBRA6-2-E |
GABA Receptor Alpha-6 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
780 |
0.43 |
Binding ≤ 10μM
|
|
GBRB2-1-E |
GABA Receptor Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
780 |
0.43 |
Binding ≤ 10μM
|
|
GBRB3-1-E |
GABA Receptor Beta-3 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
850 |
0.42 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
780 |
0.43 |
Binding ≤ 10μM
|
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 8), Other |
Other |
116 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.89 |
8.45 |
-15.54 |
0 |
4 |
0 |
43 |
278.237 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
2 |
0.49 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
2 |
0.49 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
6 |
0.46 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
2 |
0.49 |
Binding ≤ 10μM
|
|
GBRA5-1-E |
GABA Receptor Alpha-5 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
2 |
0.49 |
Binding ≤ 10μM
|
|
GBRA6-2-E |
GABA Receptor Alpha-6 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
603 |
0.35 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2 |
0.49 |
Binding ≤ 10μM
|
|
GBRB2-1-E |
GABA Receptor Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
603 |
0.35 |
Binding ≤ 10μM
|
|
GBRB3-1-E |
GABA Receptor Beta-3 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2 |
0.49 |
Binding ≤ 10μM
|
|
GBRD-1-E |
GABA Receptor Delta Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2 |
0.49 |
Binding ≤ 10μM
|
|
GBRE-1-E |
GABA Receptor Epsilon Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2 |
0.49 |
Binding ≤ 10μM
|
|
GBRG1-1-E |
GABA Receptor Gamma-1 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
2 |
0.49 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
603 |
0.35 |
Binding ≤ 10μM
|
|
GBRG3-4-E |
GABA Receptor Gamma-3 Subunit (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
2 |
0.49 |
Binding ≤ 10μM
|
|
GBRR1-1-E |
GABA Receptor Rho-1 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.49 |
Binding ≤ 10μM
|
|
Q91ZM7-1-E |
GABA Receptor Theta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2 |
0.49 |
Binding ≤ 10μM
|
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 8), Other |
Other |
2 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.24 |
0.19 |
-20.56 |
0 |
7 |
0 |
78 |
335.367 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.00 |
0.4 |
-40.43 |
1 |
7 |
1 |
80 |
336.375 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 17 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
106 |
0.44 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
6 |
0.52 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
8 |
0.52 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
2 |
0.55 |
Binding ≤ 10μM
|
|
GBRA5-1-E |
GABA Receptor Alpha-5 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRA6-2-E |
GABA Receptor Alpha-6 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
148 |
0.43 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRB2-1-E |
GABA Receptor Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
94 |
0.45 |
Binding ≤ 10μM
|
|
GBRB3-1-E |
GABA Receptor Beta-3 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
148 |
0.43 |
Binding ≤ 10μM
|
|
GBRD-1-E |
GABA Receptor Delta Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRE-1-E |
GABA Receptor Epsilon Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRG1-1-E |
GABA Receptor Gamma-1 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRG3-2-E |
GABA Receptor Gamma-3 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRP-1-E |
GABA Receptor Pi Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRT-2-E |
GABA Receptor Theta Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 8), Other |
Other |
58 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.86 |
8.07 |
-13.1 |
0 |
6 |
0 |
64 |
303.293 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
50 |
0.44 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
737 |
0.37 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
975 |
0.37 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
76 |
0.43 |
Binding ≤ 10μM
|
|
GBRA5-1-E |
GABA Receptor Alpha-5 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
35 |
0.45 |
Binding ≤ 10μM
|
|
GBRA6-2-E |
GABA Receptor Alpha-6 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
980 |
0.37 |
Binding ≤ 10μM
|
|
GBRB2-1-E |
GABA Receptor Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
980 |
0.37 |
Binding ≤ 10μM
|
|
GBRB3-1-E |
GABA Receptor Beta-3 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
383 |
0.39 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
980 |
0.37 |
Binding ≤ 10μM
|
|
GBRG3-2-E |
GABA Receptor Gamma-3 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
383 |
0.39 |
Binding ≤ 10μM
|
|
TSPO-1-E |
Peripheral-type Benzodiazepine Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4700 |
0.32 |
Binding ≤ 10μM
|
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 8), Other |
Other |
48 |
0.45 |
Binding ≤ 10μM
|
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 2), Other |
Other |
48 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.15 |
10.38 |
-16.93 |
0 |
4 |
0 |
38 |
307.397 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.15 |
10.86 |
-27.89 |
1 |
4 |
1 |
39 |
308.405 |
3 |
↓
|
|
|
|
Analogs
-
3872568
-
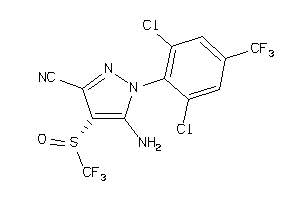
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
GBRA5-1-E |
GABA Receptor Alpha-5 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
GBRA6-2-E |
GABA Receptor Alpha-6 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
GBRB2-1-E |
GABA Receptor Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
GBRB3-1-E |
GABA Receptor Beta-3 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
28 |
0.41 |
Binding ≤ 10μM
|
|
GBRD-1-E |
GABA Receptor Delta Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
GBRE-1-E |
GABA Receptor Epsilon Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
GBRG1-7-E |
GABA Receptor Gamma-1 Subunit (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
GBRG3-2-E |
GABA Receptor Gamma-3 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
GBRP-1-E |
GABA Receptor Pi Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
GBRT-2-E |
GABA Receptor Theta Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.11 |
5.93 |
-18.04 |
2 |
5 |
0 |
85 |
437.152 |
4 |
↓
|
|
|
|
Analogs
-
1532157
-
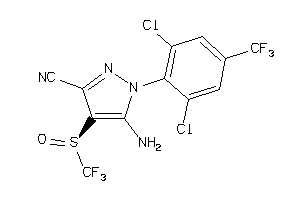
Draw
Identity
99%
90%
80%
70%
Vendors
And 11 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
GBRA5-1-E |
GABA Receptor Alpha-5 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
GBRA6-2-E |
GABA Receptor Alpha-6 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
GBRB2-1-E |
GABA Receptor Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
GBRB3-1-E |
GABA Receptor Beta-3 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
28 |
0.41 |
Binding ≤ 10μM
|
|
GBRD-1-E |
GABA Receptor Delta Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
GBRE-1-E |
GABA Receptor Epsilon Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
GBRG1-7-E |
GABA Receptor Gamma-1 Subunit (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
GBRG3-2-E |
GABA Receptor Gamma-3 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
GBRP-1-E |
GABA Receptor Pi Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
|
GBRT-2-E |
GABA Receptor Theta Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.11 |
6.32 |
-7.82 |
2 |
5 |
0 |
85 |
437.152 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
26 |
0.48 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
27 |
0.48 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
13 |
0.50 |
Binding ≤ 10μM
|
|
GBRA5-1-E |
GABA Receptor Alpha-5 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRA6-2-E |
GABA Receptor Alpha-6 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
22 |
0.49 |
Binding ≤ 10μM
|
|
GBRB3-1-E |
GABA Receptor Beta-3 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
27 |
0.48 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
27 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.51 |
8.63 |
-12.59 |
0 |
6 |
0 |
64 |
364.199 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
680 |
0.38 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
680 |
0.38 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
680 |
0.38 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
680 |
0.38 |
Binding ≤ 10μM
|
|
GBRA5-6-E |
GABA Receptor Alpha-5 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
680 |
0.38 |
Binding ≤ 10μM
|
|
GBRA6-2-E |
GABA Receptor Alpha-6 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
680 |
0.38 |
Binding ≤ 10μM
|
|
GBRB1-3-E |
GABA Receptor Beta-1 Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
680 |
0.38 |
Binding ≤ 10μM
|
|
GBRB2-1-E |
GABA Receptor Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
680 |
0.38 |
Binding ≤ 10μM
|
|
GBRB3-1-E |
GABA Receptor Beta-3 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
680 |
0.38 |
Binding ≤ 10μM
|
|
GBRD-1-E |
GABA Receptor Delta Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
680 |
0.38 |
Binding ≤ 10μM
|
|
GBRE-1-E |
GABA Receptor Epsilon Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
680 |
0.38 |
Binding ≤ 10μM
|
|
GBRG1-5-E |
GABA Receptor Gamma-1 Subunit (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
680 |
0.38 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
680 |
0.38 |
Binding ≤ 10μM
|
|
GBRG3-2-E |
GABA Receptor Gamma-3 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
680 |
0.38 |
Binding ≤ 10μM
|
|
GBRP-1-E |
GABA Receptor Pi Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
680 |
0.38 |
Binding ≤ 10μM
|
|
GBRT-2-E |
GABA Receptor Theta Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
680 |
0.38 |
Binding ≤ 10μM
|
|
Z104301-4-O |
GABA-A Receptor; Anion Channel (cluster #4 Of 8), Other |
Other |
590 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.39 |
7.6 |
-50.35 |
2 |
4 |
0 |
74 |
335.762 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.39 |
9.38 |
-52.73 |
1 |
4 |
-1 |
77 |
334.754 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.39 |
7.37 |
-46.73 |
1 |
4 |
-1 |
73 |
334.754 |
6 |
↓
|
|
|
|
Analogs
-
34533945
-
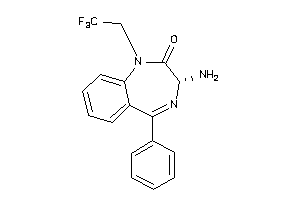
-
4375653
-
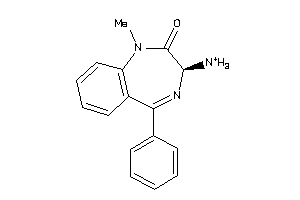
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
8500 |
0.35 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
8500 |
0.35 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
8500 |
0.35 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
8500 |
0.35 |
Binding ≤ 10μM
|
|
GBRA5-6-E |
GABA Receptor Alpha-5 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
8500 |
0.35 |
Binding ≤ 10μM
|
|
GBRA6-2-E |
GABA Receptor Alpha-6 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
8500 |
0.35 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
8500 |
0.35 |
Binding ≤ 10μM
|
|
GBRB2-1-E |
GABA Receptor Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
8500 |
0.35 |
Binding ≤ 10μM
|
|
GBRB3-1-E |
GABA Receptor Beta-3 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
8500 |
0.35 |
Binding ≤ 10μM
|
|
GBRD-1-E |
GABA Receptor Delta Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8500 |
0.35 |
Binding ≤ 10μM
|
|
GBRE-1-E |
GABA Receptor Epsilon Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
8500 |
0.35 |
Binding ≤ 10μM
|
|
GBRG1-7-E |
GABA Receptor Gamma-1 Subunit (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
8500 |
0.35 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
8500 |
0.35 |
Binding ≤ 10μM
|
|
GBRG3-2-E |
GABA Receptor Gamma-3 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
8500 |
0.35 |
Binding ≤ 10μM
|
|
GBRP-1-E |
GABA Receptor Pi Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
8500 |
0.35 |
Binding ≤ 10μM
|
|
GBRT-2-E |
GABA Receptor Theta Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
8500 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.47 |
-3.62 |
-37.81 |
3 |
4 |
1 |
60 |
266.324 |
1 |
↓
|
|
|
|
Analogs
-
6582775
-
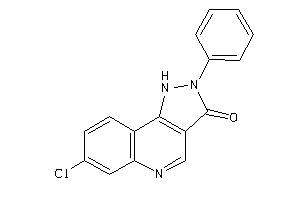
-
13448368
-
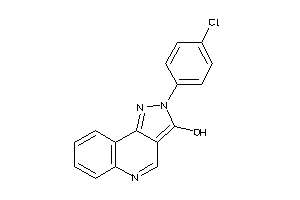
-
20744554
-
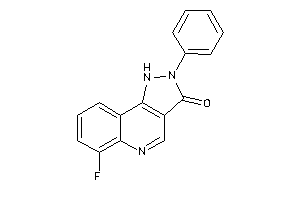
-
26507437
-
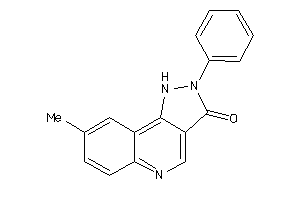
-
26507550
-
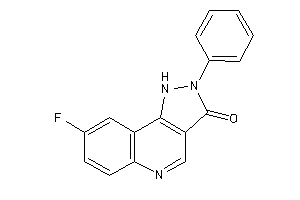
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
400 |
0.45 |
Binding ≤ 10μM
|
|
GBRA5-6-E |
GABA Receptor Alpha-5 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
200 |
0.47 |
Binding ≤ 10μM
|
|
GBRA6-2-E |
GABA Receptor Alpha-6 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
17 |
0.54 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
400 |
0.45 |
Binding ≤ 10μM
|
|
GBRB3-1-E |
GABA Receptor Beta-3 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
17 |
0.54 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
400 |
0.45 |
Binding ≤ 10μM
|
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 8), Other |
Other |
46 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.85 |
7.11 |
-8.83 |
0 |
4 |
0 |
45 |
261.284 |
1 |
↓
|
|
Ref
Reference (pH 7)
|
2.85 |
6.99 |
-12.47 |
1 |
4 |
0 |
51 |
261.284 |
1 |
↓
|
|
|
|
Analogs
-
34327160
-
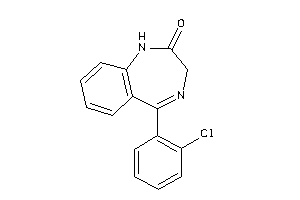
-
39296227
-
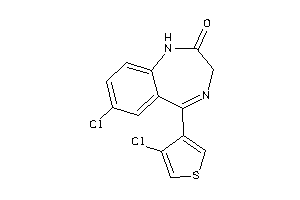
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRA5-6-E |
GABA Receptor Alpha-5 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRA6-2-E |
GABA Receptor Alpha-6 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRB2-1-E |
GABA Receptor Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRB3-1-E |
GABA Receptor Beta-3 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRD-1-E |
GABA Receptor Delta Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRE-1-E |
GABA Receptor Epsilon Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRG1-1-E |
GABA Receptor Gamma-1 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRG3-2-E |
GABA Receptor Gamma-3 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRP-1-E |
GABA Receptor Pi Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRT-2-E |
GABA Receptor Theta Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.49 |
5.58 |
-8.46 |
1 |
3 |
0 |
41 |
305.164 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.49 |
6.5 |
-34.12 |
2 |
3 |
1 |
43 |
306.172 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.93 |
-0.28 |
-16.89 |
0 |
7 |
0 |
77 |
357.801 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.16 |
-0.06 |
-43.64 |
1 |
7 |
1 |
78 |
358.809 |
1 |
↓
|
|
|
|
Analogs
-
11616881
-
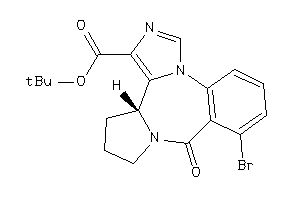
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.88 |
10.5 |
-15.44 |
0 |
6 |
0 |
64 |
418.291 |
3 |
↓
|
|
|
|
Analogs
-
4214169
-
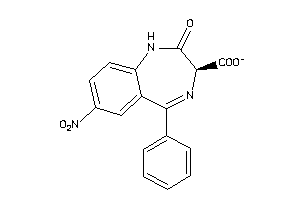
-
5850731
-
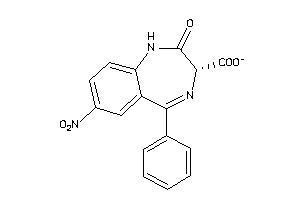
-
6095266
-
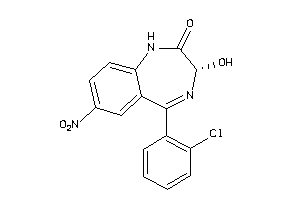
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRA5-6-E |
GABA Receptor Alpha-5 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRA6-2-E |
GABA Receptor Alpha-6 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRB2-1-E |
GABA Receptor Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRB3-1-E |
GABA Receptor Beta-3 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRD-1-E |
GABA Receptor Delta Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRE-1-E |
GABA Receptor Epsilon Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRG1-1-E |
GABA Receptor Gamma-1 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRG3-2-E |
GABA Receptor Gamma-3 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRP-1-E |
GABA Receptor Pi Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
GBRT-2-E |
GABA Receptor Theta Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
1 |
0.57 |
Binding ≤ 10μM
|
|
Z104301-1-O |
GABA-A Receptor; Anion Channel (cluster #1 Of 8), Other |
Other |
2 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.77 |
-2.23 |
-10.75 |
1 |
6 |
0 |
87 |
315.716 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
49 |
0.41 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
27 |
0.42 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
28 |
0.42 |
Binding ≤ 10μM
|
|
GBRA5-1-E |
GABA Receptor Alpha-5 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
GBRA6-2-E |
GABA Receptor Alpha-6 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
83 |
0.40 |
Binding ≤ 10μM
|
|
GBRB3-1-E |
GABA Receptor Beta-3 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
83 |
0.40 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
83 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.34 |
8.16 |
-15.95 |
0 |
7 |
0 |
74 |
341.367 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.75 |
5.73 |
-11.61 |
0 |
9 |
0 |
114 |
326.316 |
4 |
↓
|
|