|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HS90A-2-E |
Heat Shock Protein HSP 90-alpha (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3320 |
0.31 |
Binding ≤ 10μM |
|
KCMA1-1-E |
Calcium-activated Potassium Channel Subunit Alpha-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5129 |
0.30 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7943 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.31 |
6.12 |
-11.71 |
2 |
4 |
0 |
58 |
362.229 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.31 |
6.89 |
-42.7 |
1 |
4 |
-1 |
61 |
361.221 |
3 |
↓
|
|
|
|
Analogs
-
18042311
-
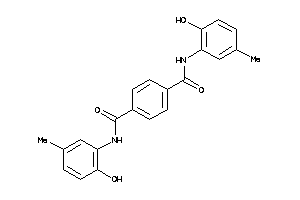
-
383803
-
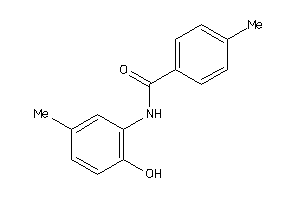
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCMA1-1-E |
Calcium-activated Potassium Channel Subunit Alpha-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9333 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.99 |
4.73 |
-13.84 |
2 |
3 |
0 |
49 |
227.263 |
2 |
↓
|
|
|
|
Analogs
-
21815380
-
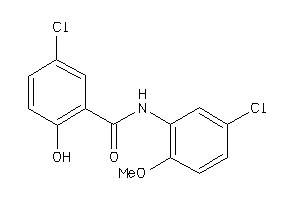
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCMA1-1-E |
Calcium-activated Potassium Channel Subunit Alpha-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.43 |
5.25 |
-10.44 |
2 |
4 |
0 |
59 |
312.152 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.43 |
6.01 |
-44.05 |
1 |
4 |
-1 |
61 |
311.144 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCMA1-1-E |
Calcium-activated Potassium Channel Subunit Alpha-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
28 |
0.53 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.20 |
5.71 |
-14.16 |
2 |
4 |
0 |
59 |
291.734 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCMA1-1-E |
Calcium-activated Potassium Channel Subunit Alpha-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
447 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.26 |
5 |
-20.99 |
2 |
7 |
0 |
104 |
288.259 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.26 |
6.02 |
-66.25 |
1 |
7 |
-1 |
107 |
287.251 |
4 |
↓
|
|
|
|
Analogs
-
21815380
-

Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCMA1-1-E |
Calcium-activated Potassium Channel Subunit Alpha-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
372 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.36 |
3.23 |
-9.17 |
3 |
4 |
0 |
70 |
298.125 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.36 |
4.23 |
-48.24 |
2 |
4 |
-1 |
72 |
297.117 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCMA1-1-E |
Calcium-activated Potassium Channel Subunit Alpha-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
251 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.63 |
0.45 |
-12.94 |
2 |
6 |
0 |
95 |
272.26 |
3 |
↓
|
|
|
|
Analogs
-
37062035
-
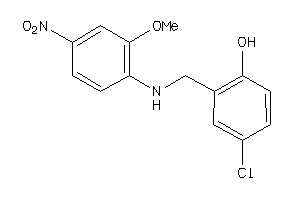
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCMA1-1-E |
Calcium-activated Potassium Channel Subunit Alpha-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
692 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.91 |
-0.3 |
-9.47 |
2 |
7 |
0 |
104 |
322.704 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCMA1-1-E |
Calcium-activated Potassium Channel Subunit Alpha-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1549 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.77 |
0.19 |
-11.55 |
1 |
5 |
0 |
68 |
239.234 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCMA1-1-E |
Calcium-activated Potassium Channel Subunit Alpha-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3090 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.23 |
0.18 |
-11.84 |
2 |
6 |
0 |
95 |
258.233 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCMA1-1-E |
Calcium-activated Potassium Channel Subunit Alpha-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3090 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.15 |
5.65 |
-12.4 |
2 |
4 |
0 |
59 |
291.734 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.15 |
6.4 |
-46.36 |
1 |
4 |
-1 |
61 |
290.726 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCMA1-1-E |
Calcium-activated Potassium Channel Subunit Alpha-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2512 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.65 |
0.44 |
-10.55 |
2 |
6 |
0 |
95 |
272.26 |
3 |
↓
|
|
|
|
Analogs
-
5915889
-
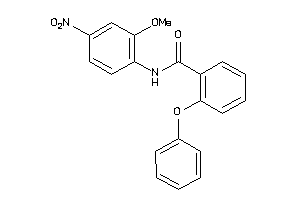
-
5916419
-
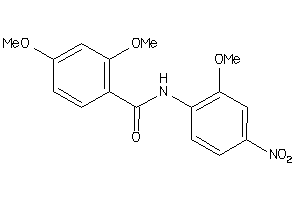
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCMA1-1-E |
Calcium-activated Potassium Channel Subunit Alpha-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
191 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.26 |
5.07 |
-11.06 |
2 |
7 |
0 |
104 |
288.259 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.26 |
6.08 |
-50.21 |
1 |
7 |
-1 |
107 |
287.251 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCMA1-1-E |
Calcium-activated Potassium Channel Subunit Alpha-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3467 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.10 |
2.28 |
-8.73 |
2 |
6 |
0 |
95 |
326.23 |
4 |
↓
|
|