|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 25 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RY2-1-E |
Purinergic Receptor P2Y2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
130 |
0.33 |
Functional ≤ 10μM
|
|
P2RY4-1-E |
Pyrimidinergic Receptor P2Y4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
90 |
0.34 |
Functional ≤ 10μM
|
|
P2RY6-1-E |
Pyrimidinergic Receptor P2Y6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
-
12959016
-
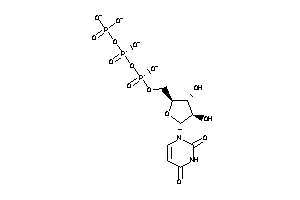
Draw
Identity
99%
90%
80%
70%
Vendors
And 27 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.57 |
-4.59 |
-370.18 |
3 |
17 |
-4 |
276 |
480.108 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.57 |
-5.75 |
-234.35 |
4 |
17 |
-3 |
273 |
481.116 |
8 |
↓
|
|
|
|
Analogs
-
15637617
-
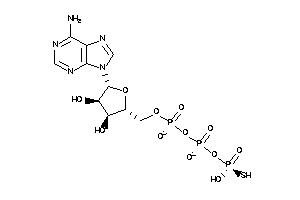
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RX1-1-E |
P2X Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2300 |
0.25 |
Functional ≤ 10μM
|
|
P2RX2-1-E |
P2X Purinoceptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.26 |
Functional ≤ 10μM
|
|
P2RX3-1-E |
P2X Purinoceptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
690 |
0.28 |
Functional ≤ 10μM
|
|
P2RX4-1-E |
P2X Purinoceptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2300 |
0.25 |
Functional ≤ 10μM
|
|
P2RX5-1-E |
P2X Purinoceptor 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
600 |
0.28 |
Functional ≤ 10μM
|
|
P2RX6-1-E |
P2X Purinoceptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.27 |
Functional ≤ 10μM
|
|
P2RY1-1-E |
P2Y Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.27 |
Functional ≤ 10μM
|
|
P2RY2-1-E |
Purinergic Receptor P2Y2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1720 |
0.26 |
Functional ≤ 10μM
|
|
P2Y11-1-E |
Purinergic Receptor P2Y11 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.25 |
Functional ≤ 10μM
|
|
P2Y12-1-E |
Purinergic Receptor P2Y12 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1300 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.75 |
-2.04 |
-153.5 |
6 |
17 |
-2 |
268 |
521.234 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.92 |
-1.22 |
-126.98 |
6 |
17 |
-1 |
266 |
522.242 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.92 |
-5.6 |
-128.24 |
6 |
17 |
-1 |
266 |
522.242 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 45 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KAP2-1-E |
CAMP-dependent Protein Kinase Type II-alpha Regulatory Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.23 |
Binding ≤ 10μM
|
|
KAPCA-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.23 |
Binding ≤ 10μM
|
|
KAPCB-1-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.23 |
Binding ≤ 10μM
|
|
MAPK2-1-E |
MAP Kinase-activated Protein Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6300 |
0.23 |
Binding ≤ 10μM
|
|
P2RY1-1-E |
Purinergic Receptor P2Y1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2800 |
0.25 |
Binding ≤ 10μM
|
|
P2RX1-1-E |
P2X Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
56 |
0.33 |
Functional ≤ 10μM
|
|
P2RX2-1-E |
P2X Purinoceptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.26 |
Functional ≤ 10μM
|
|
P2RX3-1-E |
P2X Purinoceptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
340 |
0.29 |
Functional ≤ 10μM
|
|
P2RX4-1-E |
P2X Purinoceptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.28 |
Functional ≤ 10μM
|
|
P2RX5-1-E |
P2X Purinoceptor 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.27 |
Functional ≤ 10μM
|
|
P2RX6-1-E |
P2X Purinoceptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.27 |
Functional ≤ 10μM
|
|
P2RY1-1-E |
P2Y Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7200 |
0.23 |
Functional ≤ 10μM
|
|
P2RY2-1-E |
Purinergic Receptor P2Y2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
96 |
0.32 |
Functional ≤ 10μM
|
|
P2RY4-1-E |
Pyrimidinergic Receptor P2Y4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1800 |
0.26 |
Functional ≤ 10μM
|
|
P2Y11-1-E |
Purinergic Receptor P2Y11 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3300 |
0.25 |
Functional ≤ 10μM |
|
P2Y12-1-E |
Purinergic Receptor P2Y12 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3190 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
1-Amino-4-(2'-(4",6"-dichloro-s-triazin-2-yl)amino)phenylamino)9,10-dihydro-9,10-dioxoanthracene-2,4'-disulphonic acid; 2-Anthracenesulfonic acid, 1-amino-4-((3-((4,6-dichloro-1,3,5-triazin-2-yl)amino)-4-sulfophenyl)amino)-9,10-dihydro-9,10-dioxo-; 2-Anth
1-Amino-4-(2'-(4",6"-dichloro-s-…
Find On:
PubMed —
Wikipedia —
Google
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RX1-1-E |
P2X Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
422 |
0.22 |
Functional ≤ 10μM |
|
P2RX2-1-E |
P2X Purinoceptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
79 |
0.24 |
Functional ≤ 10μM |
|
P2RX3-1-E |
P2X Purinoceptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
494 |
0.22 |
Functional ≤ 10μM |
|
P2RY2-1-E |
Purinergic Receptor P2Y2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.17 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.56 |
4.68 |
-94.13 |
4 |
14 |
-2 |
237 |
635.423 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RY2-1-E |
Purinergic Receptor P2Y2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.35 |
Functional ≤ 10μM
|
|
P2RY4-1-E |
Pyrimidinergic Receptor P2Y4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
350 |
0.31 |
Functional ≤ 10μM
|
|
P2RY6-1-E |
Pyrimidinergic Receptor P2Y6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RY2-1-E |
Purinergic Receptor P2Y2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.35 |
Functional ≤ 10μM
|
|
P2RY4-1-E |
Pyrimidinergic Receptor P2Y4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
350 |
0.31 |
Functional ≤ 10μM
|
|
P2RY6-1-E |
Pyrimidinergic Receptor P2Y6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.36 |
-3.29 |
-374.05 |
3 |
16 |
-4 |
259 |
496.176 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.36 |
-4.44 |
-237.97 |
4 |
16 |
-3 |
256 |
497.184 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RY2-1-E |
Purinergic Receptor P2Y2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.35 |
Functional ≤ 10μM
|
|
P2RY4-1-E |
Pyrimidinergic Receptor P2Y4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
350 |
0.31 |
Functional ≤ 10μM
|
|
P2RY6-1-E |
Pyrimidinergic Receptor P2Y6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RY2-1-E |
Purinergic Receptor P2Y2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
50 |
0.35 |
Functional ≤ 10μM
|
|
P2RY4-1-E |
Pyrimidinergic Receptor P2Y4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
350 |
0.31 |
Functional ≤ 10μM
|
|
P2RY6-1-E |
Pyrimidinergic Receptor P2Y6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5NTD-1-E |
5'-nucleotidase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4810 |
0.26 |
Binding ≤ 10μM
|
|
P2Y12-1-E |
Purinergic Receptor P2Y12 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7350 |
0.25 |
Binding ≤ 10μM
|
|
P2RY2-1-E |
Purinergic Receptor P2Y2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5310 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.65 |
5.16 |
-43.59 |
3 |
7 |
-1 |
129 |
427.845 |
3 |
↓
|
|
|
|
Analogs
-
31351672
-
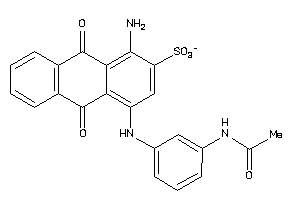
-
31476925
-
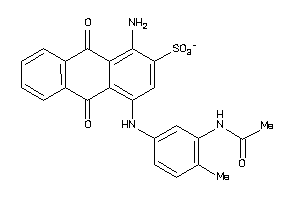
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RX2-1-E |
P2X Purinoceptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
690 |
0.27 |
Functional ≤ 10μM |
|
P2RY2-1-E |
Purinergic Receptor P2Y2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5810 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.19 |
3.51 |
-50.69 |
4 |
9 |
-1 |
158 |
450.452 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RY2-1-E |
Purinergic Receptor P2Y2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.19 |
Functional ≤ 10μM
|
|
P2RY6-1-E |
Pyrimidinergic Receptor P2Y6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1600 |
0.23 |
Functional ≤ 10μM
|
|
P2Y14-1-E |
P2Y Purinoceptor 14 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.03 |
-14.08 |
-161 |
7 |
19 |
-2 |
303 |
564.286 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5NTD-1-E |
5'-nucleotidase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
530 |
0.27 |
Binding ≤ 10μM
|
|
Q80Z26-1-E |
Nucleoside Triphosphate Diphosphohydrolase 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1500 |
0.25 |
Binding ≤ 10μM
|
|
P2RY2-1-E |
Purinergic Receptor P2Y2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6670 |
0.23 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.13 |
6.88 |
-47.05 |
3 |
7 |
-1 |
129 |
443.46 |
3 |
↓
|
|