|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
0.64 |
-10.62 |
0 |
3 |
0 |
43 |
247.253 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
6700 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
6.84 |
-17.42 |
2 |
5 |
0 |
71 |
405.932 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
7400 |
0.22 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.20 |
-3.93 |
-17.61 |
2 |
6 |
0 |
80 |
498.029 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 14 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.81 |
7.33 |
-10.83 |
1 |
4 |
0 |
54 |
272.311 |
2 |
↓
|
|
|
|
Analogs
-
6582528
-
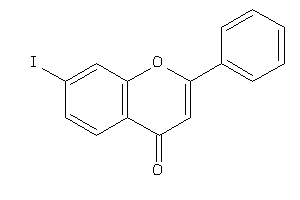
-
39119560
-
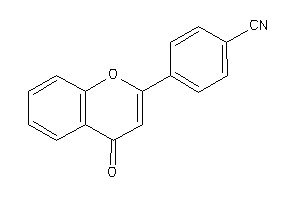
-
38933
-
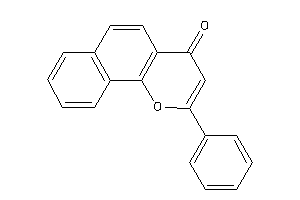
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.90 |
2.24 |
-12.55 |
0 |
2 |
0 |
30 |
272.303 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP3A4-3-E |
Cytochrome P450 3A4 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
700 |
0.20 |
ADME/T ≤ 10μM
|
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
155 |
0.22 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.49 |
-2.13 |
-19.91 |
7 |
13 |
0 |
217 |
594.525 |
8 |
↓
|
|
|
|
Analogs
-
4517652
-
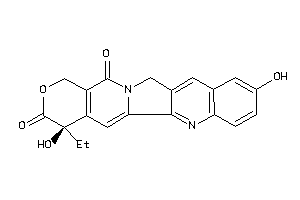
-
4823972
-
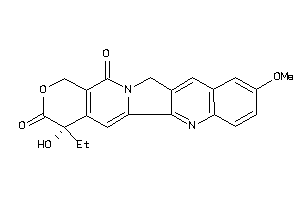
Draw
Identity
99%
90%
80%
70%
Vendors
And 35 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
110 |
0.36 |
Binding ≤ 10μM
|
|
Z80099-1-O |
COLO 205 (Colon Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
9 |
0.42 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
410 |
0.33 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
9 |
0.42 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
4100 |
0.28 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
18 |
0.40 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
12 |
0.41 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
110 |
0.36 |
Functional ≤ 10μM
|
|
Z80697-1-O |
Bel-7402 (Hepatoma Cells) (cluster #1 Of 3), Other |
Other |
280 |
0.34 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
148 |
0.35 |
Functional ≤ 10μM
|
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
981 |
0.31 |
Functional ≤ 10μM
|
|
Z81034-1-O |
A2780 (Ovarian Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
700 |
0.32 |
Functional ≤ 10μM
|
|
Z81072-1-O |
Jurkat (Acute Leukemic T-cells) (cluster #1 Of 10), Other |
Other |
30 |
0.39 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
829 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.53 |
3.91 |
-13.24 |
2 |
7 |
0 |
102 |
364.357 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.28 |
3.5 |
-56.32 |
2 |
7 |
0 |
102 |
364.357 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.28 |
4.82 |
-55.03 |
2 |
7 |
0 |
104 |
364.357 |
1 |
↓
|
|
|
|
Analogs
-
598071
-
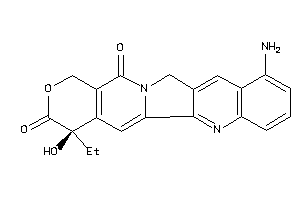
Draw
Identity
99%
90%
80%
70%
Vendors
And 7 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
840 |
0.32 |
Binding ≤ 10μM
|
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4200 |
0.28 |
Binding ≤ 10μM |
|
Z80099-1-O |
COLO 205 (Colon Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
7 |
0.42 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
7 |
0.42 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
13 |
0.41 |
Functional ≤ 10μM |
|
Z80898-1-O |
NCI-H928 (cluster #1 Of 1), Other |
Other |
8 |
0.42 |
Functional ≤ 10μM
|
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
4 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.77 |
4.71 |
-13.32 |
3 |
7 |
0 |
107 |
363.373 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.30 |
Binding ≤ 10μM
|
|
ABL2-1-E |
Tyrosine-protein Kinase ABL2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.35 |
Binding ≤ 10μM
|
|
ACK1-1-E |
Tyrosine Kinase Non-receptor Protein 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.33 |
Binding ≤ 10μM
|
|
ALK-1-E |
ALK Tyrosine Kinase Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.26 |
Binding ≤ 10μM
|
|
BCR-1-E |
Breakpoint Cluster Region Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.30 |
Binding ≤ 10μM
|
|
BLK-1-E |
Tyrosine-protein Kinase BLK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.32 |
Binding ≤ 10μM
|
|
BMX-1-E |
Tyrosine-protein Kinase BMX (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.31 |
Binding ≤ 10μM
|
|
BTK-1-E |
Tyrosine-protein Kinase BTK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.33 |
Binding ≤ 10μM
|
|
CSK-1-E |
Tyrosine-protein Kinase CSK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
63 |
0.28 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
53 |
0.28 |
Binding ≤ 10μM
|
|
EPHB2-1-E |
Ephrin Type-B Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.31 |
Binding ≤ 10μM
|
|
EPHB4-1-E |
Ephrin Type-B Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.32 |
Binding ≤ 10μM
|
|
FAK1-1-E |
Focal Adhesion Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
538 |
0.24 |
Binding ≤ 10μM
|
|
FAK2-1-E |
Protein-tyrosine Kinase 2-beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
134 |
0.27 |
Binding ≤ 10μM
|
|
FER-1-E |
Tyrosine-protein Kinase FER (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
490 |
0.25 |
Binding ≤ 10μM
|
|
FES-1-E |
Tyrosine-protein Kinase FES (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
358 |
0.25 |
Binding ≤ 10μM
|
|
FGR-1-E |
Tyrosine-protein Kinase FGR (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.35 |
Binding ≤ 10μM
|
|
FRK-1-E |
Tyrosine-protein Kinase FRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.34 |
Binding ≤ 10μM
|
|
FYN-1-E |
Tyrosine-protein Kinase FYN (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2 |
0.34 |
Binding ≤ 10μM
|
|
HCK-1-E |
Tyrosine-protein Kinase HCK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.33 |
Binding ≤ 10μM
|
|
IRAK4-1-E |
Interleukin-1 Receptor-associated Kinase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
574 |
0.24 |
Binding ≤ 10μM
|
|
KCC1D-1-E |
CaM Kinase I Delta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
92 |
0.27 |
Binding ≤ 10μM
|
|
KCC2G-1-E |
CaM Kinase II Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
184 |
0.26 |
Binding ≤ 10μM
|
|
KIT-1-E |
Stem Cell Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6313 |
0.20 |
Binding ≤ 10μM
|
|
KSYK-1-E |
Tyrosine-protein Kinase SYK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
52 |
0.28 |
Binding ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.35 |
Binding ≤ 10μM
|
|
LYN-1-E |
Tyrosine-protein Kinase Lyn (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.31 |
Binding ≤ 10μM
|
|
M4K2-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10 |
0.31 |
Binding ≤ 10μM
|
|
M4K5-1-E |
Mitogen-activated Protein Kinase Kinase Kinase Kinase 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
MINK1-1-E |
Misshapen-like Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
22 |
0.30 |
Binding ≤ 10μM
|
|
NPM-1-E |
Nucleophosmin (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
200 |
0.26 |
Binding ≤ 10μM
|
|
NTRK1-1-E |
Nerve Growth Factor Receptor Trk-A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
22 |
0.30 |
Binding ≤ 10μM
|
|
NTRK2-1-E |
Neurotrophic Tyrosine Kinase Receptor Type 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
27 |
0.29 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.33 |
Binding ≤ 10μM
|
|
STK10-1-E |
Serine/threonine-protein Kinase 10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
52 |
0.28 |
Binding ≤ 10μM
|
|
STK24-1-E |
Serine/threonine-protein Kinase 24 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.33 |
Binding ≤ 10μM
|
|
STK4-1-E |
Serine/threonine-protein Kinase MST1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
191 |
0.26 |
Binding ≤ 10μM
|
|
TBK1-1-E |
Serine/threonine-protein Kinase TBK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
471 |
0.25 |
Binding ≤ 10μM
|
|
TEC-1-E |
Tyrosine-protein Kinase TEC (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
282 |
0.25 |
Binding ≤ 10μM
|
|
TIE2-1-E |
Tyrosine-protein Kinase TIE-2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1191 |
0.23 |
Binding ≤ 10μM
|
|
TXK-1-E |
Tyrosine-protein Kinase TXK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
40 |
0.29 |
Binding ≤ 10μM
|
|
UFO-1-E |
Tyrosine-protein Kinase Receptor UFO (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
174 |
0.26 |
Binding ≤ 10μM
|
|
YES-1-E |
Tyrosine-protein Kinase YES (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
490 |
0.25 |
Functional ≤ 10μM
|
|
ABL2-1-E |
Tyrosine-protein Kinase ABL2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.35 |
Functional ≤ 10μM
|
|
ERBB2-2-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2600 |
0.22 |
Functional ≤ 10μM
|
|
FYN-1-E |
Tyrosine-protein Kinase FYN (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
410 |
0.25 |
Functional ≤ 10μM
|
|
PGFRA-1-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.21 |
Functional ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.21 |
Functional ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.27 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
100 |
0.27 |
Functional ≤ 10μM
|
|
Z80036-2-O |
BaF3 (IL-3-dependent Pro-B-cells) (cluster #2 Of 3), Other |
Other |
200 |
0.26 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
5000 |
0.21 |
Functional ≤ 10μM
|
|
Z80186-6-O |
K562 (Erythroleukemia Cells) (cluster #6 Of 11), Other |
Other |
20 |
0.30 |
Functional ≤ 10μM
|
|
Z80416-1-O |
Rat1 (Fibroblast Cells) (cluster #1 Of 1), Other |
Other |
490 |
0.25 |
Functional ≤ 10μM
|
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
3910 |
0.21 |
Functional ≤ 10μM
|
|
Z81125-1-O |
KU812 Cell Line (cluster #1 Of 1), Other |
Other |
5 |
0.32 |
Functional ≤ 10μM
|
|
Z81328-1-O |
Src-transformed (Fibroblast Cells) (cluster #1 Of 1), Other |
Other |
100 |
0.27 |
Functional ≤ 10μM
|
|
ABL-1-V |
Tyrosine-protein Kinase V-ABL (cluster #1 Of 1), Viral |
Viruses |
1 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.98 |
9.49 |
-44.8 |
2 |
8 |
1 |
84 |
531.464 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.98 |
7.02 |
-12.44 |
1 |
8 |
0 |
83 |
530.456 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.98 |
9.35 |
-46.97 |
2 |
8 |
1 |
84 |
531.464 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.35 |
Binding ≤ 10μM
|
|
TOP2B-1-E |
DNA Topoisomerase II Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.35 |
Binding ≤ 10μM
|
|
Z100745-2-O |
MX1 (Breast Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
4 |
0.49 |
Functional ≤ 10μM
|
|
Z80063-1-O |
CCRF S-180 (cluster #1 Of 1), Other |
Other |
30 |
0.44 |
Functional ≤ 10μM
|
|
Z80064-8-O |
CCRF-CEM (T-cell Leukemia) (cluster #8 Of 9), Other |
Other |
753 |
0.36 |
Functional ≤ 10μM
|
|
Z80084-1-O |
CEM-VLB (Vinblastine-resistant T-cell Leukemia Cells) (cluster #1 Of 2), Other |
Other |
3790 |
0.32 |
Functional ≤ 10μM
|
|
Z80085-1-O |
CEM-VM1 (Etoposide-resistant T-cell Leukemia Cells) (cluster #1 Of 2), Other |
Other |
1600 |
0.34 |
Functional ≤ 10μM
|
|
Z80117-2-O |
DC3F/AD-II (cluster #2 Of 2), Other |
Other |
54 |
0.42 |
Functional ≤ 10μM
|
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
25 |
0.44 |
Functional ≤ 10μM
|
|
Z80166-4-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #4 Of 12), Other |
Other |
880 |
0.35 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
16 |
0.45 |
Functional ≤ 10μM
|
|
Z80555-1-O |
TSGH (cluster #1 Of 1), Other |
Other |
450 |
0.37 |
Functional ≤ 10μM
|
|
Z80682-7-O |
A549 (Lung Carcinoma Cells) (cluster #7 Of 11), Other |
Other |
47 |
0.43 |
Functional ≤ 10μM
|
|
Z80967-1-O |
Hone-1 Cell Line (cluster #1 Of 2), Other |
Other |
310 |
0.38 |
Functional ≤ 10μM
|
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
1400 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.03 |
-4.69 |
-10.2 |
4 |
4 |
0 |
71 |
315.376 |
3 |
↓
|
|
|
|
Analogs
-
8582318
-
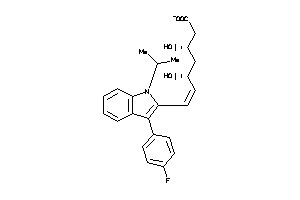
-
8582319
-
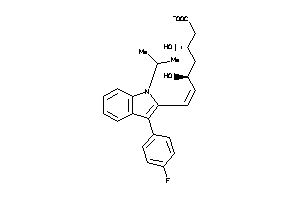
-
1530639
-
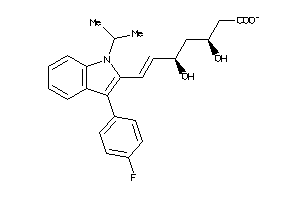
Draw
Identity
99%
90%
80%
70%
Vendors
And 25 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.14 |
11.25 |
-51.38 |
2 |
5 |
-1 |
86 |
410.465 |
8 |
↓
|
|
|
|
Analogs
-
22065493
-
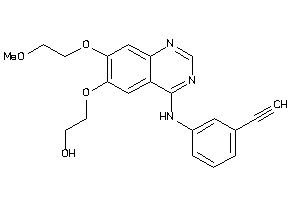
Draw
Identity
99%
90%
80%
70%
Vendors
And 64 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AAK1-1-E |
Adaptor-associated Kinase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4400 |
0.26 |
Binding ≤ 10μM
|
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
770 |
0.30 |
Binding ≤ 10μM
|
|
ABL2-1-E |
Tyrosine-protein Kinase ABL2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
300 |
0.31 |
Binding ≤ 10μM
|
|
ADCK3-1-E |
Chaperone Activity Of Bc1 Complex-like, Mitochondrial (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.28 |
Binding ≤ 10μM
|
|
ADCK4-1-E |
Uncharacterized AarF Domain-containing Protein Kinase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2500 |
0.27 |
Binding ≤ 10μM
|
|
ALK-1-E |
ALK Tyrosine Kinase Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.29 |
Binding ≤ 10μM
|
|
AURKA-3-E |
Serine/threonine-protein Kinase Aurora-A (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
3800 |
0.26 |
Binding ≤ 10μM
|
|
AURKB-1-E |
Serine/threonine-protein Kinase Aurora-B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.28 |
Binding ≤ 10μM
|
|
AURKC-1-E |
Serine/threonine-protein Kinase Aurora-C (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
600 |
0.30 |
Binding ≤ 10μM
|
|
BLK-1-E |
Tyrosine-protein Kinase BLK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
190 |
0.32 |
Binding ≤ 10μM
|
|
BMP2K-1-E |
BMP-2-inducible Protein Kinase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1800 |
0.28 |
Binding ≤ 10μM
|
|
CTRO-1-E |
Citron Rho-interacting Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
680 |
0.30 |
Binding ≤ 10μM
|
|
DDR1-1-E |
Epithelial Discoidin Domain-containing Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
790 |
0.29 |
Binding ≤ 10μM
|
|
DMPK-1-E |
Myotonin-protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2900 |
0.27 |
Binding ≤ 10μM
|
|
E2AK2-1-E |
Interferon-induced, Double-stranded RNA-activated Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1300 |
0.28 |
Binding ≤ 10μM
|
|
E2AK4-1-E |
Eukaryotic Translation Initiation Factor 2-alpha Kinase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4400 |
0.26 |
Binding ≤ 10μM
|
|
EGFR-3-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
51 |
0.35 |
Binding ≤ 10μM
|
|
EPHA3-1-E |
Ephrin Type-A Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2400 |
0.27 |
Binding ≤ 10μM
|
|
EPHA6-1-E |
Ephrin Type-A Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
930 |
0.29 |
Binding ≤ 10μM
|
|
EPHA7-1-E |
Ephrin Type-A Receptor 7 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.28 |
Binding ≤ 10μM
|
|
EPHA8-1-E |
Ephrin Type-A Receptor 8 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
940 |
0.29 |
Binding ≤ 10μM
|
|
ERBB2-2-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
870 |
0.29 |
Binding ≤ 10μM
|
|
ERBB4-1-E |
Receptor Protein-tyrosine Kinase ErbB-4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1130 |
0.29 |
Binding ≤ 10μM
|
|
FGFR2-1-E |
Fibroblast Growth Factor Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.28 |
Binding ≤ 10μM
|
|
FGR-1-E |
Tyrosine-protein Kinase FGR (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1100 |
0.29 |
Binding ≤ 10μM
|
|
FLT3-1-E |
Tyrosine-protein Kinase Receptor FLT3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
820 |
0.29 |
Binding ≤ 10μM
|
|
FRK-1-E |
Tyrosine-protein Kinase FRK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.28 |
Binding ≤ 10μM
|
|
GAK-1-E |
Serine/threonine-protein Kinase GAK (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
40 |
0.36 |
Binding ≤ 10μM
|
|
HCK-1-E |
Tyrosine-protein Kinase HCK (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1800 |
0.28 |
Binding ≤ 10μM
|
|
JAK3-2-E |
Tyrosine-protein Kinase JAK3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
930 |
0.29 |
Binding ≤ 10μM
|
|
KC1D-1-E |
Casein Kinase I Delta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3500 |
0.26 |
Binding ≤ 10μM
|
|
KIT-1-E |
Stem Cell Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3100 |
0.27 |
Binding ≤ 10μM
|
|
KS6A4-1-E |
Ribosomal Protein S6 Kinase Alpha 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7000 |
0.25 |
Binding ≤ 10μM
|
|
LCK-1-E |
Tyrosine-protein Kinase LCK (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
530 |
0.30 |
Binding ≤ 10μM
|
|
LTK-1-E |
Leukocyte Tyrosine Kinase Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
890 |
0.29 |
Binding ≤ 10μM
|
|
LYN-1-E |
Tyrosine-protein Kinase Lyn (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
530 |
0.30 |
Binding ≤ 10μM
|
|
MET-1-E |
Hepatocyte Growth Factor Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3800 |
0.26 |
Binding ≤ 10μM
|
|
MK04-1-E |
Mitogen-activated Protein Kinase 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2500 |
0.27 |
Binding ≤ 10μM
|
|
MK09-1-E |
C-Jun N-terminal Kinase 2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4000 |
0.26 |
Binding ≤ 10μM
|
|
MKNK1-1-E |
MAP Kinase-interacting Serine/threonine-protein Kinase MNK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
690 |
0.30 |
Binding ≤ 10μM
|
|
MKNK2-1-E |
MAP Kinase Signal-integrating Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.28 |
Binding ≤ 10μM
|
|
MRCKG-1-E |
Serine/threonine-protein Kinase MRCK Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3400 |
0.26 |
Binding ≤ 10μM
|
|
MYLK2-1-E |
Myosin Light Chain Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
970 |
0.29 |
Binding ≤ 10μM
|
|
PGFRA-1-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1800 |
0.28 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.28 |
Binding ≤ 10μM
|
|
RET-1-E |
Tyrosine-protein Kinase Receptor RET (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
330 |
0.31 |
Binding ≤ 10μM
|
|
RIPK2-1-E |
Serine/threonine-protein Kinase RIPK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
680 |
0.30 |
Binding ≤ 10μM
|
|
SLK-1-E |
Serine/threonine-protein Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
26 |
0.37 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
700 |
0.30 |
Binding ≤ 10μM
|
|
STK10-1-E |
Serine/threonine-protein Kinase 10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
83 |
0.34 |
Binding ≤ 10μM
|
|
TBA1A-2-E |
Tubulin Alpha-3 Chain (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
230 |
0.32 |
Binding ≤ 10μM
|
|
TIE1-1-E |
Tyrosine-protein Kinase Receptor Tie-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
850 |
0.29 |
Binding ≤ 10μM
|
|
TNI3K-1-E |
Serine/threonine-protein Kinase TNNI3K (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
570 |
0.30 |
Binding ≤ 10μM
|
|
TNK1-1-E |
Non-receptor Tyrosine-protein Kinase TNK1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
630 |
0.30 |
Binding ≤ 10μM
|
|
TYRO3-1-E |
Tyrosine-protein Kinase Receptor TYRO3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3900 |
0.26 |
Binding ≤ 10μM
|
|
ULK3-1-E |
Serine/threonine-protein Kinase ULK3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
630 |
0.30 |
Binding ≤ 10μM
|
|
VGFR1-1-E |
Vascular Endothelial Growth Factor Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4400 |
0.26 |
Binding ≤ 10μM
|
|
VGFR3-1-E |
Vascular Endothelial Growth Factor Receptor 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2100 |
0.27 |
Binding ≤ 10μM
|
|
YES-1-E |
Tyrosine-protein Kinase YES (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2200 |
0.27 |
Binding ≤ 10μM
|
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
19 |
0.37 |
Functional ≤ 10μM
|
|
ERBB2-2-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
299 |
0.31 |
Functional ≤ 10μM
|
|
Z103205-1-O |
A431 (cluster #1 Of 4), Other |
Other |
96 |
0.34 |
Functional ≤ 10μM
|
|
Z103348-1-O |
NCI-H2122 (cluster #1 Of 2), Other |
Other |
1000 |
0.29 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
20 |
0.37 |
Functional ≤ 10μM |
|
Z80322-2-O |
NCI-H358 (Lung Carcinama Cells) (cluster #2 Of 2), Other |
Other |
6000 |
0.25 |
Functional ≤ 10μM
|
|
Z80475-3-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #3 Of 3), Other |
Other |
1560 |
0.28 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
270 |
0.32 |
Functional ≤ 10μM |
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
80 |
0.34 |
Functional ≤ 10μM |
|
Z81072-6-O |
Jurkat (Acute Leukemic T-cells) (cluster #6 Of 10), Other |
Other |
2000 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.80 |
9.59 |
-15.77 |
1 |
7 |
0 |
75 |
393.443 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-1-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.28 |
Binding ≤ 10μM
|
|
ERBB2-2-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.28 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8500 |
0.18 |
Binding ≤ 10μM
|
|
RIPK2-1-E |
Serine/threonine-protein Kinase RIPK2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3600 |
0.19 |
Binding ≤ 10μM
|
|
SLK-1-E |
Serine/threonine-protein Kinase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9300 |
0.18 |
Binding ≤ 10μM
|
|
STK10-1-E |
Serine/threonine-protein Kinase 10 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4400 |
0.19 |
Binding ≤ 10μM
|
|
TBA1A-1-E |
Tubulin Alpha-3 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
54 |
0.25 |
Binding ≤ 10μM
|
|
Z103205-1-O |
A431 (cluster #1 Of 4), Other |
Other |
970 |
0.21 |
Functional ≤ 10μM
|
|
Z80046-1-O |
BT-474 (Breast Ductal Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
80 |
0.25 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
6600 |
0.18 |
Functional ≤ 10μM
|
|
Z80300-1-O |
N87 (Gastric Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
80 |
0.25 |
Functional ≤ 10μM
|
|
Z80475-3-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #3 Of 3), Other |
Other |
4350 |
0.19 |
Functional ≤ 10μM
|
|
Z80493-1-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
3 |
0.30 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
8500 |
0.18 |
Functional ≤ 10μM
|
|
Z80954-1-O |
HFF (Foreskin Fibroblasts) (cluster #1 Of 4), Other |
Other |
9900 |
0.18 |
Functional ≤ 10μM
|
|
Z80961-1-O |
HN5 (Squamous Cell Carcinoma) (cluster #1 Of 1), Other |
Other |
25 |
0.27 |
Functional ≤ 10μM
|
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
6270 |
0.18 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
5900 |
0.18 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
5400 |
0.18 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.16 |
-3.53 |
-72.53 |
3 |
8 |
1 |
110 |
582.077 |
11 |
↓
|
|
|
|
Analogs
-
5733913
-
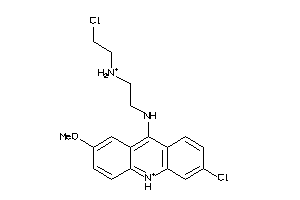
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
280 |
0.44 |
Functional ≤ 10μM
|
|
Z80186-6-O |
K562 (Erythroleukemia Cells) (cluster #6 Of 11), Other |
Other |
7000 |
0.34 |
Functional ≤ 10μM |
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
9300 |
0.34 |
Functional ≤ 10μM |
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
6000 |
0.35 |
Functional ≤ 10μM |
|
Z81252-10-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #10 Of 11), Other |
Other |
8120 |
0.34 |
Functional ≤ 10μM
|
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 3), Other |
Other |
2070 |
0.38 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.05 |
-4.51 |
-93.85 |
5 |
4 |
2 |
63 |
303.793 |
4 |
↓
|
|
|
|
Analogs
-
33822027
-
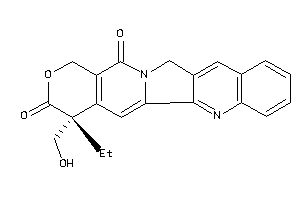
-
33822028
-
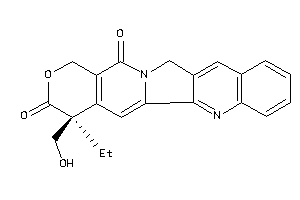
-
1087
-
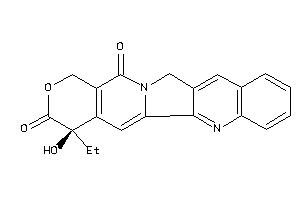
Draw
Identity
99%
90%
80%
70%
Vendors
And 55 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.31 |
Binding ≤ 10μM
|
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9000 |
0.27 |
Binding ≤ 10μM
|
|
CHK1-1-E |
Serine/threonine-protein Kinase Chk1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Functional ≤ 10μM
|
|
SSR1-1-E |
Somatostatin Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Functional ≤ 10μM
|
|
SSR2-1-E |
Somatostatin Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Functional ≤ 10μM
|
|
SSR3-1-E |
Somatostatin Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Functional ≤ 10μM
|
|
SSR4-1-E |
Somatostatin Receptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Functional ≤ 10μM
|
|
SSR5-1-E |
Somatostatin Receptor 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.47 |
Functional ≤ 10μM
|
|
TOP1-1-E |
DNA Topoisomerase I (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
33 |
0.40 |
Functional ≤ 10μM
|
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
33 |
0.40 |
Functional ≤ 10μM
|
|
TOP2B-1-E |
DNA Topoisomerase II Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
33 |
0.40 |
Functional ≤ 10μM
|
|
Z100501-1-O |
Lu1 (Lung Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
29 |
0.41 |
Functional ≤ 10μM
|
|
Z100733-1-O |
CT26 (Colon Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
34 |
0.40 |
Functional ≤ 10μM
|
|
Z100752-1-O |
Renca (Renal Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
304 |
0.35 |
Functional ≤ 10μM
|
|
Z103205-1-O |
A431 (cluster #1 Of 4), Other |
Other |
11 |
0.43 |
Functional ≤ 10μM
|
|
Z103408-1-O |
SW626 (cluster #1 Of 2), Other |
Other |
29 |
0.41 |
Functional ≤ 10μM
|
|
Z103413-1-O |
SW948 (cluster #1 Of 1), Other |
Other |
160 |
0.37 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
794 |
0.33 |
Functional ≤ 10μM
|
|
Z80012-1-O |
A 172 (Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
29 |
0.41 |
Functional ≤ 10μM
|
|
Z80012-1-O |
A 172 (Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
140 |
0.37 |
Functional ≤ 10μM |
|
Z80019-1-O |
A-427 (Lung Carcinoma Cells) (cluster #1 Of 4), Other |
Other |
24 |
0.41 |
Functional ≤ 10μM
|
|
Z80026-1-O |
AGS (Gastric Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
20 |
0.41 |
Functional ≤ 10μM
|
|
Z80054-1-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
9550 |
0.27 |
Functional ≤ 10μM |
|
Z80056-1-O |
CAKI-2 (Renal Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
3960 |
0.29 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
5300 |
0.28 |
Functional ≤ 10μM
|
|
Z80068-3-O |
CCRF-SB (Lymphoblastic Leukemia Cells) (cluster #3 Of 5), Other |
Other |
3 |
0.46 |
Functional ≤ 10μM
|
|
Z80099-1-O |
COLO 205 (Colon Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
5 |
0.45 |
Functional ≤ 10μM
|
|
Z80115-1-O |
DC3F (cluster #1 Of 1), Other |
Other |
6 |
0.44 |
Functional ≤ 10μM |
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
102 |
0.38 |
Functional ≤ 10μM
|
|
Z80120-1-O |
DLD-1 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
210 |
0.36 |
Functional ≤ 10μM |
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
940 |
0.32 |
Functional ≤ 10μM
|
|
Z80145-3-O |
H69 (cluster #3 Of 4), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z80152-1-O |
HCT-8 (Ileocecal Adenocarcinoma) (cluster #1 Of 2), Other |
Other |
7 |
0.44 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
89 |
0.38 |
Functional ≤ 10μM
|
|
Z80164-1-O |
HT-1080 (Fibrosarcoma Cells) (cluster #1 Of 6), Other |
Other |
80 |
0.38 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
90 |
0.38 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
80 |
0.38 |
Functional ≤ 10μM
|
|
Z80188-1-O |
KB 3-1 (Cervical Epithelial Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
20 |
0.41 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
35 |
0.40 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
60 |
0.39 |
Functional ≤ 10μM
|
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
3100 |
0.30 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
7420 |
0.28 |
Functional ≤ 10μM
|
|
Z80269-1-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
17 |
0.42 |
Functional ≤ 10μM
|
|
Z80295-2-O |
MT4 (Lymphocytes) (cluster #2 Of 8), Other |
Other |
4 |
0.45 |
Functional ≤ 10μM
|
|
Z80316-1-O |
NCI-H128 (cluster #1 Of 1), Other |
Other |
18 |
0.42 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
9 |
0.43 |
Functional ≤ 10μM
|
|
Z80372-1-O |
P388CPT5 (cluster #1 Of 1), Other |
Other |
2800 |
0.30 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
57 |
0.39 |
Functional ≤ 10μM
|
|
Z80414-1-O |
Raji (B-lymphoblastic Cells) (cluster #1 Of 3), Other |
Other |
1000 |
0.32 |
Functional ≤ 10μM
|
|
Z80466-1-O |
SF268 (cluster #1 Of 4), Other |
Other |
40 |
0.40 |
Functional ≤ 10μM |
|
Z80475-1-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #1 Of 3), Other |
Other |
20 |
0.41 |
Functional ≤ 10μM
|
|
Z80482-2-O |
SK-MEL-2 (Melanoma Cells) (cluster #2 Of 4), Other |
Other |
7860 |
0.27 |
Functional ≤ 10μM |
|
Z80485-1-O |
SK-MEL-28 (Melanoma Cells) (cluster #1 Of 6), Other |
Other |
40 |
0.40 |
Functional ≤ 10μM
|
|
Z80493-1-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
51 |
0.39 |
Functional ≤ 10μM
|
|
Z80495-1-O |
SK-VLB (cluster #1 Of 1), Other |
Other |
53 |
0.39 |
Functional ≤ 10μM
|
|
Z80532-1-O |
T-24 (Bladder Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
88 |
0.38 |
Functional ≤ 10μM
|
|
Z80548-1-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #1 Of 5), Other |
Other |
600 |
0.34 |
Functional ≤ 10μM
|
|
Z80623-1-O |
Panel (56 Tumour Cell Lines) (cluster #1 Of 1), Other |
Other |
40 |
0.40 |
Functional ≤ 10μM
|
|
Z80647-1-O |
833K Cell Line (cluster #1 Of 1), Other |
Other |
10 |
0.43 |
Functional ≤ 10μM |
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
91 |
0.38 |
Functional ≤ 10μM |
|
Z80697-1-O |
Bel-7402 (Hepatoma Cells) (cluster #1 Of 3), Other |
Other |
7 |
0.44 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
207 |
0.36 |
Functional ≤ 10μM
|
|
Z80751-1-O |
Caov-3 Cell Line (cluster #1 Of 1), Other |
Other |
30 |
0.41 |
Functional ≤ 10μM |
|
Z80751-1-O |
Caov-3 Cell Line (cluster #1 Of 1), Other |
Other |
32 |
0.40 |
Functional ≤ 10μM
|
|
Z80784-1-O |
Col2 (Colon Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
45 |
0.40 |
Functional ≤ 10μM
|
|
Z80799-1-O |
RPMI 8402 (Pre-T-lymphoblastoid Cells) (cluster #1 Of 1), Other |
Other |
6 |
0.44 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
1 |
0.48 |
Functional ≤ 10μM
|
|
Z80898-1-O |
NCI-H928 (cluster #1 Of 1), Other |
Other |
8 |
0.44 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
9 |
0.43 |
Functional ≤ 10μM
|
|
Z80930-1-O |
HEC-1B Cell Line (cluster #1 Of 1), Other |
Other |
60 |
0.39 |
Functional ≤ 10μM
|
|
Z80930-1-O |
HEC-1B Cell Line (cluster #1 Of 1), Other |
Other |
8640 |
0.27 |
Functional ≤ 10μM
|
|
Z81017-1-O |
WiDr (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
17 |
0.42 |
Functional ≤ 10μM
|
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
60 |
0.39 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
330 |
0.35 |
Functional ≤ 10μM
|
|
Z81034-1-O |
A2780 (Ovarian Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
7 |
0.44 |
Functional ≤ 10μM
|
|
Z81054-1-O |
SF-268 (Glioblastoma Cells) (cluster #1 Of 3), Other |
Other |
59 |
0.39 |
Functional ≤ 10μM
|
|
Z81072-1-O |
Jurkat (Acute Leukemic T-cells) (cluster #1 Of 10), Other |
Other |
6 |
0.44 |
Functional ≤ 10μM
|
|
Z81080-1-O |
KATO-III Cell Line (cluster #1 Of 1), Other |
Other |
448 |
0.34 |
Functional ≤ 10μM
|
|
Z81080-1-O |
KATO-III Cell Line (cluster #1 Of 1), Other |
Other |
1170 |
0.32 |
Functional ≤ 10μM |
|
Z81084-1-O |
KB VCR R (cluster #1 Of 1), Other |
Other |
9 |
0.43 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
9 |
0.43 |
Functional ≤ 10μM
|
|
Z81134-1-O |
L2987 (Lung Adenocarcinoma Cells) (cluster #1 Of 1), Other |
Other |
400 |
0.34 |
Functional ≤ 10μM
|
|
Z81160-1-O |
Lewis Lung Carcinoma Cell Line (cluster #1 Of 1), Other |
Other |
33 |
0.40 |
Functional ≤ 10μM
|
|
Z81164-1-O |
LL Cell Line (cluster #1 Of 2), Other |
Other |
33 |
0.40 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
86 |
0.38 |
Functional ≤ 10μM
|
|
Z81184-1-O |
LOX IMVI (Melanoma Cells) (cluster #1 Of 5), Other |
Other |
48 |
0.39 |
Functional ≤ 10μM
|
|
Z81245-1-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
710 |
0.33 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
940 |
0.32 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
970 |
0.32 |
Functional ≤ 10μM
|
|
Z81331-1-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
2 |
0.47 |
Functional ≤ 10μM
|
|
Z81335-1-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
83 |
0.38 |
Functional ≤ 10μM
|
|
Z80291-2-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
890 |
0.33 |
ADME/T ≤ 10μM |
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
500 |
0.34 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.03 |
6.76 |
-12.81 |
1 |
6 |
0 |
81 |
348.358 |
1 |
↓
|
|
|
|
Analogs
-
35572125
-
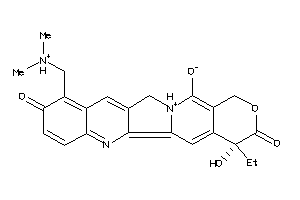
-
35643838
-
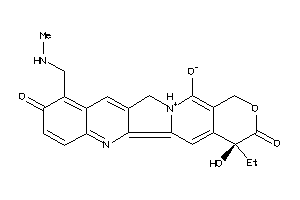
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HIF1A-1-E |
Hypoxia-inducible Factor 1-alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
60 |
0.33 |
Binding ≤ 10μM
|
|
TOP1-1-E |
DNA Topoisomerase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.33 |
Binding ≤ 10μM
|
|
Z100519-1-O |
KETR3 (Renal Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
5 |
0.37 |
Functional ≤ 10μM
|
|
Z103203-1-O |
A375 (cluster #1 Of 3), Other |
Other |
13 |
0.36 |
Functional ≤ 10μM
|
|
Z80019-1-O |
A-427 (Lung Carcinoma Cells) (cluster #1 Of 4), Other |
Other |
49 |
0.33 |
Functional ≤ 10μM
|
|
Z80026-1-O |
AGS (Gastric Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
300 |
0.29 |
Functional ≤ 10μM
|
|
Z80039-1-O |
BGC-823 (Stomach Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
3800 |
0.24 |
Functional ≤ 10μM
|
|
Z80105-1-O |
COR-L23 (Lung Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
20 |
0.35 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
1200 |
0.27 |
Functional ≤ 10μM
|
|
Z80145-3-O |
H69 (cluster #3 Of 4), Other |
Other |
63 |
0.33 |
Functional ≤ 10μM
|
|
Z80152-1-O |
HCT-8 (Ileocecal Adenocarcinoma) (cluster #1 Of 2), Other |
Other |
74 |
0.32 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
52 |
0.33 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
880 |
0.27 |
Functional ≤ 10μM
|
|
Z80186-1-O |
K562 (Erythroleukemia Cells) (cluster #1 Of 11), Other |
Other |
1700 |
0.26 |
Functional ≤ 10μM
|
|
Z80188-1-O |
KB 3-1 (Cervical Epithelial Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
440 |
0.29 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
601 |
0.28 |
Functional ≤ 10μM |
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
36 |
0.34 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
5550 |
0.24 |
Functional ≤ 10μM
|
|
Z80269-1-O |
MKN-45 (Gastric Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
38 |
0.34 |
Functional ≤ 10μM
|
|
Z80316-1-O |
NCI-H128 (cluster #1 Of 1), Other |
Other |
31 |
0.34 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
45 |
0.33 |
Functional ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
45 |
0.33 |
Functional ≤ 10μM
|
|
Z80475-1-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #1 Of 3), Other |
Other |
26 |
0.34 |
Functional ≤ 10μM
|
|
Z80482-2-O |
SK-MEL-2 (Melanoma Cells) (cluster #2 Of 4), Other |
Other |
140 |
0.31 |
Functional ≤ 10μM
|
|
Z80493-1-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
48 |
0.33 |
Functional ≤ 10μM
|
|
Z80495-1-O |
SK-VLB (cluster #1 Of 1), Other |
Other |
150 |
0.31 |
Functional ≤ 10μM
|
|
Z80548-1-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #1 Of 5), Other |
Other |
1500 |
0.26 |
Functional ≤ 10μM
|
|
Z80559-1-O |
U251 (cluster #1 Of 3), Other |
Other |
500 |
0.28 |
Functional ≤ 10μM
|
|
Z80596-1-O |
WISH (Amniotic Epithelial Cells) (cluster #1 Of 1), Other |
Other |
1300 |
0.27 |
Functional ≤ 10μM
|
|
Z80623-1-O |
Panel (56 Tumour Cell Lines) (cluster #1 Of 1), Other |
Other |
160 |
0.31 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
87 |
0.32 |
Functional ≤ 10μM
|
|
Z80697-1-O |
Bel-7402 (Hepatoma Cells) (cluster #1 Of 3), Other |
Other |
78 |
0.32 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
104 |
0.32 |
Functional ≤ 10μM
|
|
Z80799-1-O |
RPMI 8402 (Pre-T-lymphoblastoid Cells) (cluster #1 Of 1), Other |
Other |
21 |
0.35 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
1100 |
0.27 |
Functional ≤ 10μM
|
|
Z81017-1-O |
WiDr (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
56 |
0.33 |
Functional ≤ 10μM
|
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
60 |
0.33 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
7100 |
0.23 |
Functional ≤ 10μM
|
|
Z81034-1-O |
A2780 (Ovarian Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
8700 |
0.23 |
Functional ≤ 10μM
|
|
Z81084-1-O |
KB VCR R (cluster #1 Of 1), Other |
Other |
339 |
0.29 |
Functional ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 6), Other |
Other |
70 |
0.32 |
Functional ≤ 10μM
|
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 5), Other |
Other |
9 |
0.36 |
Functional ≤ 10μM
|
|
Z81184-1-O |
LOX IMVI (Melanoma Cells) (cluster #1 Of 5), Other |
Other |
5 |
0.37 |
Functional ≤ 10μM
|
|
Z81245-1-O |
MDA-MB-435 (Breast Carcinoma Cells) (cluster #1 Of 6), Other |
Other |
79 |
0.32 |
Functional ≤ 10μM
|
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
1600 |
0.26 |
Functional ≤ 10μM
|
|
Z81248-1-O |
Malme-3M (Melanoma Cells) (cluster #1 Of 3), Other |
Other |
1000 |
0.27 |
Functional ≤ 10μM
|
|
Z81331-1-O |
SW-620 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
3 |
0.38 |
Functional ≤ 10μM
|
|
Z81342-1-O |
Tumour Cell Lines (cluster #1 Of 1), Other |
Other |
40 |
0.33 |
Functional ≤ 10μM
|
|
Z80682-2-O |
A549 (Lung Carcinoma Cells) (cluster #2 Of 3), Other |
Other |
3100 |
0.25 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.31 |
6.39 |
-40.99 |
3 |
8 |
0 |
108 |
422.461 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.31 |
4.23 |
-51.04 |
2 |
8 |
0 |
107 |
421.453 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.31 |
2.6 |
-55.53 |
2 |
8 |
0 |
105 |
421.453 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
5550 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.27 |
-0.06 |
-14.08 |
3 |
6 |
0 |
100 |
292.287 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
53 |
0.27 |
Binding ≤ 10μM |
|
Z80026-1-O |
AGS (Gastric Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
6700 |
0.19 |
Functional ≤ 10μM |
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
3330 |
0.20 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
7020 |
0.19 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
940 |
0.22 |
Functional ≤ 10μM
|
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
2100 |
0.21 |
Functional ≤ 10μM
|
|
Z81034-3-O |
A2780 (Ovarian Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
3000 |
0.20 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.94 |
2.26 |
-55.93 |
1 |
6 |
1 |
59 |
596.977 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
12 |
0.29 |
Binding ≤ 10μM
|
|
KIF11-1-E |
Kinesin-like Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
53 |
0.27 |
Binding ≤ 10μM |
|
Z80026-1-O |
AGS (Gastric Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
6700 |
0.19 |
Functional ≤ 10μM |
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
3330 |
0.20 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
7020 |
0.19 |
Functional ≤ 10μM
|
|
Z80928-3-O |
HCT-116 (Colon Carcinoma Cells) (cluster #3 Of 9), Other |
Other |
940 |
0.22 |
Functional ≤ 10μM
|
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
2100 |
0.21 |
Functional ≤ 10μM
|
|
Z81034-3-O |
A2780 (Ovarian Carcinoma Cells) (cluster #3 Of 10), Other |
Other |
3000 |
0.20 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.94 |
17.46 |
-55.26 |
1 |
6 |
1 |
60 |
596.977 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
700 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.48 |
-3.9 |
-14.3 |
4 |
4 |
0 |
82 |
287.319 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.48 |
-3.91 |
-31.9 |
5 |
4 |
1 |
83 |
288.327 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
4760 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.29 |
0.82 |
-10.99 |
4 |
5 |
0 |
95 |
270.317 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.29 |
1.16 |
-36.89 |
5 |
5 |
1 |
96 |
271.325 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
1970 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.51 |
0.73 |
-10.33 |
4 |
5 |
0 |
95 |
259.29 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.51 |
1.02 |
-32.55 |
5 |
5 |
1 |
96 |
260.298 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.85 |
7.77 |
-13.28 |
3 |
5 |
0 |
77 |
423.925 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.85 |
8.09 |
-33.86 |
4 |
5 |
1 |
78 |
424.933 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81020-7-O |
HepG2 (Hepatoblastoma Cells) (cluster #7 Of 8), Other |
Other |
19 |
0.51 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.42 |
1.92 |
-11.35 |
4 |
5 |
0 |
91 |
299.355 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.42 |
2.12 |
-36.04 |
5 |
5 |
1 |
92 |
300.363 |
3 |
↓
|
|