|
|
|
|
|
Analogs
-
4015531
-
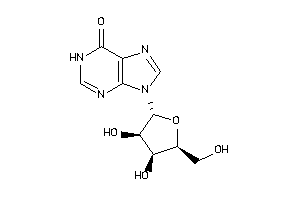
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80583-7-O |
Vero (Kidney Cells) (cluster #7 Of 7), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.82 |
-4.43 |
-23.87 |
4 |
9 |
0 |
133 |
268.229 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.36 |
-6.52 |
-59.54 |
3 |
9 |
-1 |
137 |
267.221 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.51 |
2.96 |
-10.94 |
0 |
4 |
0 |
44 |
138.174 |
0 |
↓
|
|
|
|
Analogs
-
4098919
-
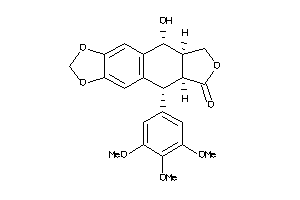
-
4098920
-
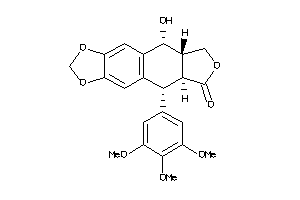
-
4475321
-
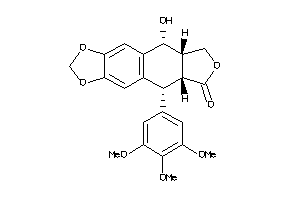
-
11682059
-
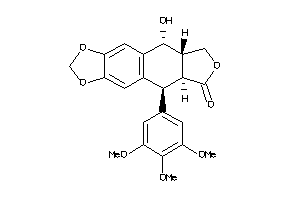
-
16969582
-
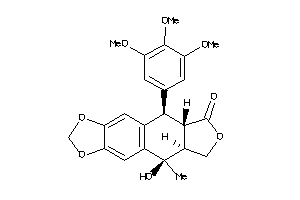
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GCR-1-E |
Glucocorticoid Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
14 |
0.37 |
Binding ≤ 10μM
|
|
O46399-1-E |
Beta Tubulin (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.26 |
Binding ≤ 10μM
|
|
Q862F3-2-E |
Tubulin Alpha Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.26 |
Binding ≤ 10μM |
|
Q862L2-2-E |
Tubulin Alpha-1 Chain (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.26 |
Binding ≤ 10μM |
|
Q9MZB0-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.26 |
Binding ≤ 10μM
|
|
TBA1A-1-E |
Tubulin Alpha-3 Chain (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
2180 |
0.26 |
Binding ≤ 10μM
|
|
TBA4A-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.27 |
Binding ≤ 10μM
|
|
TBB-1-E |
Tubulin Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1560 |
0.27 |
Binding ≤ 10μM
|
|
TBB1-1-E |
Tubulin Beta-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
460 |
0.30 |
Binding ≤ 10μM
|
|
TBB2B-1-E |
Tubulin Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2500 |
0.26 |
Binding ≤ 10μM |
|
TBB3-1-E |
Tubulin Beta-3 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2180 |
0.26 |
Binding ≤ 10μM
|
|
TBB4A-1-E |
Tubulin Beta-4 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.25 |
Binding ≤ 10μM
|
|
TBB4B-1-E |
Tubulin Beta-2 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.25 |
Binding ≤ 10μM
|
|
TBB5-1-E |
Tubulin Beta-5 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.25 |
Binding ≤ 10μM
|
|
TBB8-1-E |
Tubulin Beta-8 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4000 |
0.25 |
Binding ≤ 10μM
|
|
TBG1-1-E |
Tubulin Gamma-1 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2180 |
0.26 |
Binding ≤ 10μM
|
|
Q862F3-1-E |
Tubulin Alpha Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
590 |
0.29 |
Functional ≤ 10μM
|
|
Q862L2-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
590 |
0.29 |
Functional ≤ 10μM
|
|
TBA1A-1-E |
Tubulin Alpha-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
460 |
0.30 |
Functional ≤ 10μM
|
|
TBB1-1-E |
Tubulin Beta-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
460 |
0.30 |
Functional ≤ 10μM
|
|
TBB2B-1-E |
Tubulin Beta Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
590 |
0.29 |
Functional ≤ 10μM
|
|
Z100695-1-O |
WRL68 (Embryonic Hepatoma Cells) (cluster #1 Of 1), Other |
Other |
4800 |
0.25 |
Functional ≤ 10μM
|
|
Z101973-1-O |
Paracentrotus Lividus (cluster #1 Of 1), Other |
Other |
500 |
0.29 |
Functional ≤ 10μM
|
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
10000 |
0.23 |
Functional ≤ 10μM
|
|
Z50426-3-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #3 Of 9), Other |
Other |
12 |
0.37 |
Functional ≤ 10μM
|
|
Z50602-2-O |
Human Herpesvirus 1 (cluster #2 Of 5), Other |
Other |
50 |
0.34 |
Functional ≤ 10μM
|
|
Z50607-1-O |
Human Immunodeficiency Virus 1 (cluster #1 Of 10), Other |
Other |
3 |
0.40 |
Functional ≤ 10μM
|
|
Z50651-2-O |
Vesicular Stomatitis Virus (cluster #2 Of 2), Other |
Other |
100 |
0.33 |
Functional ≤ 10μM
|
|
Z80002-1-O |
1A9 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
3 |
0.40 |
Functional ≤ 10μM
|
|
Z80054-1-O |
Caco-2 (Colon Adenocarcinoma Cells) (cluster #1 Of 4), Other |
Other |
500 |
0.29 |
Functional ≤ 10μM
|
|
Z80110-1-O |
CV-1 (Kidney Cells) (cluster #1 Of 2), Other |
Other |
29 |
0.35 |
Functional ≤ 10μM |
|
Z80121-1-O |
DMS-79 (Small Cell Lung Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
23 |
0.36 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
52 |
0.34 |
Functional ≤ 10μM
|
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
2750 |
0.26 |
Functional ≤ 10μM
|
|
Z80164-1-O |
HT-1080 (Fibrosarcoma Cells) (cluster #1 Of 6), Other |
Other |
8100 |
0.24 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
50 |
0.34 |
Functional ≤ 10μM |
|
Z80186-6-O |
K562 (Erythroleukemia Cells) (cluster #6 Of 11), Other |
Other |
30 |
0.35 |
Functional ≤ 10μM |
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
8500 |
0.24 |
Functional ≤ 10μM
|
|
Z80284-3-O |
MOLT-3 (T-lymphoblastic Leukemia Cells) (cluster #3 Of 3), Other |
Other |
14 |
0.37 |
Functional ≤ 10μM
|
|
Z80354-4-O |
OVCAR-3 (Ovarian Adenocarcinoma Cells) (cluster #4 Of 4), Other |
Other |
460 |
0.30 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
50 |
0.34 |
Functional ≤ 10μM |
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 10), Other |
Other |
13 |
0.37 |
Functional ≤ 10μM
|
|
Z80414-1-O |
Raji (B-lymphoblastic Cells) (cluster #1 Of 3), Other |
Other |
18 |
0.36 |
Functional ≤ 10μM
|
|
Z80427-1-O |
RKO (Colon Carcinoma) (cluster #1 Of 1), Other |
Other |
29 |
0.35 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
7630 |
0.24 |
Functional ≤ 10μM
|
|
Z80697-1-O |
Bel-7402 (Hepatoma Cells) (cluster #1 Of 3), Other |
Other |
9 |
0.38 |
Functional ≤ 10μM
|
|
Z80819-1-O |
J774.2 (cluster #1 Of 1), Other |
Other |
13 |
0.37 |
Functional ≤ 10μM |
|
Z80847-1-O |
FaDu (Pharyngeal Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
7 |
0.38 |
Functional ≤ 10μM
|
|
Z80866-1-O |
GLC4 Cell Line (cluster #1 Of 1), Other |
Other |
60 |
0.34 |
Functional ≤ 10μM
|
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
7 |
0.38 |
Functional ≤ 10μM
|
|
Z80936-1-O |
HEK293 (Embryonic Kidney Fibroblasts) (cluster #1 Of 4), Other |
Other |
17 |
0.36 |
Functional ≤ 10μM
|
|
Z81020-3-O |
HepG2 (Hepatoblastoma Cells) (cluster #3 Of 8), Other |
Other |
500 |
0.29 |
Functional ≤ 10μM
|
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 6), Other |
Other |
8 |
0.38 |
Functional ≤ 10μM
|
|
Z81135-1-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #1 Of 4), Other |
Other |
150 |
0.32 |
Functional ≤ 10μM
|
|
Z81244-1-O |
J774 (Macrophage Cells) (cluster #1 Of 1), Other |
Other |
16 |
0.36 |
Functional ≤ 10μM |
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
69 |
0.33 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
8 |
0.38 |
Functional ≤ 10μM
|
|
Z81335-2-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
1000 |
0.28 |
Functional ≤ 10μM
|
|
Z80052-1-O |
C8166 (Leukemic T-cells) (cluster #1 Of 1), Other |
Other |
150 |
0.32 |
ADME/T ≤ 10μM
|
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
34 |
0.35 |
ADME/T ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 3), Other |
Other |
23 |
0.36 |
ADME/T ≤ 10μM
|
|
Z81115-2-O |
KB (Squamous Cell Carcinoma) (cluster #2 Of 3), Other |
Other |
5 |
0.39 |
ADME/T ≤ 10μM
|
|
Z81135-3-O |
L6 (Skeletal Muscle Myoblast Cells) (cluster #3 Of 6), Other |
Other |
9 |
0.38 |
ADME/T ≤ 10μM |
|
Z81170-1-O |
LNCaP (Prostate Carcinoma) (cluster #1 Of 1), Other |
Other |
31 |
0.35 |
ADME/T ≤ 10μM
|
|
VE2-1-V |
Human Papillomavirus Regulatory Protein E2 (cluster #1 Of 1), Viral |
Viruses |
1050 |
0.28 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GCR_HUMAN |
P04150
|
Glucocorticoid Receptor, Human |
14 |
0.37 |
Binding ≤ 1μM
|
|
TBA1A_PIG |
P02550
|
Tubulin Alpha Chain, Pig |
460 |
0.30 |
Binding ≤ 1μM
|
|
TBA1A_RAT |
P68370
|
Tubulin Alpha-1 Chain, Rat |
460 |
0.30 |
Binding ≤ 1μM
|
|
TBB1_HUMAN |
Q9H4B7
|
Tubulin Beta-1 Chain, Human |
1000 |
0.28 |
Binding ≤ 1μM
|
|
TBB4B_HUMAN |
P68371
|
Tubulin Beta-2 Chain, Human |
1000 |
0.28 |
Binding ≤ 1μM
|
|
TBB3_HUMAN |
Q13509
|
Tubulin Beta-3 Chain, Human |
1000 |
0.28 |
Binding ≤ 1μM
|
|
TBB4A_HUMAN |
P04350
|
Tubulin Beta-4 Chain, Human |
1000 |
0.28 |
Binding ≤ 1μM
|
|
TBB8_HUMAN |
Q3ZCM7
|
Tubulin Beta-8 Chain, Human |
1000 |
0.28 |
Binding ≤ 1μM
|
|
O46399_SHEEP |
O46399
|
Beta Tubulin, Sheep |
3000 |
0.26 |
Binding ≤ 10μM
|
|
GCR_HUMAN |
P04150
|
Glucocorticoid Receptor, Human |
14 |
0.37 |
Binding ≤ 10μM
|
|
VE2_HPV1A |
P03118
|
Regulatory Protein E2, Hpv1a |
1050 |
0.28 |
Binding ≤ 10μM
|
|
Q862F3_BOVIN |
Q862F3
|
Tubulin Alpha Chain, Bovin |
1700 |
0.27 |
Binding ≤ 10μM
|
|
TBA1A_PIG |
P02550
|
Tubulin Alpha Chain, Pig |
1560 |
0.27 |
Binding ≤ 10μM
|
|
TBA1A_RAT |
P68370
|
Tubulin Alpha-1 Chain, Rat |
460 |
0.30 |
Binding ≤ 10μM
|
|
Q862L2_BOVIN |
Q862L2
|
Tubulin Alpha-1 Chain, Bovin |
1700 |
0.27 |
Binding ≤ 10μM
|
|
TBA4A_HUMAN |
P68366
|
Tubulin Alpha-1 Chain, Human |
1700 |
0.27 |
Binding ≤ 10μM
|
|
Q9MZB0_SHEEP |
Q9MZB0
|
Tubulin Alpha-1 Chain, Sheep |
3000 |
0.26 |
Binding ≤ 10μM
|
|
TBA1A_HUMAN |
Q71U36
|
Tubulin Alpha-3 Chain, Human |
2180 |
0.26 |
Binding ≤ 10μM
|
|
TBB2B_BOVIN |
Q6B856
|
Tubulin Beta Chain, Bovin |
1700 |
0.27 |
Binding ≤ 10μM
|
|
TBB_PIG |
P02554
|
Tubulin Beta Chain, Pig |
1560 |
0.27 |
Binding ≤ 10μM
|
|
TBB2B_RAT |
Q3KRE8
|
Tubulin Beta Chain, Rat |
2180 |
0.26 |
Binding ≤ 10μM
|
|
TBB1_HUMAN |
Q9H4B7
|
Tubulin Beta-1 Chain, Human |
1000 |
0.28 |
Binding ≤ 10μM
|
|
TBB4B_HUMAN |
P68371
|
Tubulin Beta-2 Chain, Human |
1000 |
0.28 |
Binding ≤ 10μM
|
|
TBB3_RAT |
Q4QRB4
|
Tubulin Beta-3 Chain, Rat |
2180 |
0.26 |
Binding ≤ 10μM
|
|
TBB3_HUMAN |
Q13509
|
Tubulin Beta-3 Chain, Human |
1000 |
0.28 |
Binding ≤ 10μM
|
|
TBB4A_HUMAN |
P04350
|
Tubulin Beta-4 Chain, Human |
1000 |
0.28 |
Binding ≤ 10μM
|
|
TBB5_HUMAN |
P07437
|
Tubulin Beta-5 Chain, Human |
2180 |
0.26 |
Binding ≤ 10μM
|
|
TBB8_HUMAN |
Q3ZCM7
|
Tubulin Beta-8 Chain, Human |
1000 |
0.28 |
Binding ≤ 10μM
|
|
TBG1_RAT |
P83888
|
Tubulin Gamma-1 Chain, Rat |
2180 |
0.26 |
Binding ≤ 10μM
|
|
Z80002 |
Z80002
|
1A9 (Ovarian Adenocarcinoma Cells) |
3 |
0.40 |
Functional ≤ 10μM
|
|
Z80682 |
Z80682
|
A549 (Lung Carcinoma Cells) |
12 |
0.37 |
Functional ≤ 10μM
|
|
Z80697 |
Z80697
|
Bel-7402 (Hepatoma Cells) |
9.1 |
0.38 |
Functional ≤ 10μM
|
|
Z80054 |
Z80054
|
Caco-2 (Colon Adenocarcinoma Cells) |
500 |
0.29 |
Functional ≤ 10μM
|
|
Z80874 |
Z80874
|
CEM (T-cell Leukemia) |
7.2 |
0.38 |
Functional ≤ 10μM
|
|
Z80110 |
Z80110
|
CV-1 (Kidney Cells) |
29 |
0.35 |
Functional ≤ 10μM
|
|
Z80121 |
Z80121
|
DMS-79 (Small Cell Lung Carcinoma Cells) |
23 |
0.36 |
Functional ≤ 10μM
|
|
Z80125 |
Z80125
|
DU-145 (Prostate Carcinoma) |
2970 |
0.26 |
Functional ≤ 10μM
|
|
Z80847 |
Z80847
|
FaDu (Pharyngeal Carcinoma Cells) |
7.2 |
0.38 |
Functional ≤ 10μM
|
|
Z80866 |
Z80866
|
GLC4 Cell Line |
150 |
0.32 |
Functional ≤ 10μM
|
|
Z81335 |
Z81335
|
HCT-15 (Colon Adenocarcinoma Cells) |
1000 |
0.28 |
Functional ≤ 10μM
|
|
Z80936 |
Z80936
|
HEK293 (Embryonic Kidney Fibroblasts) |
17 |
0.36 |
Functional ≤ 10μM
|
|
Z81247 |
Z81247
|
HeLa (Cervical Adenocarcinoma Cells) |
100 |
0.33 |
Functional ≤ 10μM |
|
Z81020 |
Z81020
|
HepG2 (Hepatoblastoma Cells) |
24 |
0.36 |
Functional ≤ 10μM
|
|
Z80156 |
Z80156
|
HL-60 (Promyeloblast Leukemia Cells) |
2750 |
0.26 |
Functional ≤ 10μM
|
|
Z80164 |
Z80164
|
HT-1080 (Fibrosarcoma Cells) |
8100 |
0.24 |
Functional ≤ 10μM
|
|
Z80166 |
Z80166
|
HT-29 (Colon Adenocarcinoma Cells) |
11 |
0.37 |
Functional ≤ 10μM
|
|
Z50602 |
Z50602
|
Human Herpesvirus 1 |
50 |
0.34 |
Functional ≤ 10μM
|
|
Z50607 |
Z50607
|
Human Immunodeficiency Virus 1 |
2.9 |
0.40 |
Functional ≤ 10μM
|
|
Z81244 |
Z81244
|
J774 (Macrophage Cells) |
16.4 |
0.36 |
Functional ≤ 10μM
|
|
Z80819 |
Z80819
|
J774.2 |
11.3 |
0.37 |
Functional ≤ 10μM
|
|
Z80186 |
Z80186
|
K562 (Erythroleukemia Cells) |
30 |
0.35 |
Functional ≤ 10μM |
|
Z81115 |
Z81115
|
KB (Squamous Cell Carcinoma) |
0.5 |
0.43 |
Functional ≤ 10μM |
|
Z81135 |
Z81135
|
L6 (Skeletal Muscle Myoblast Cells) |
12 |
0.37 |
Functional ≤ 10μM
|
|
Z80224 |
Z80224
|
MCF7 (Breast Carcinoma Cells) |
15 |
0.37 |
Functional ≤ 10μM
|
|
Z81252 |
Z81252
|
MDA-MB-231 (Breast Adenocarcinoma Cells) |
26 |
0.35 |
Functional ≤ 10μM
|
|
Z80284 |
Z80284
|
MOLT-3 (T-lymphoblastic Leukemia Cells) |
14 |
0.37 |
Functional ≤ 10μM
|
|
Z80354 |
Z80354
|
OVCAR-3 (Ovarian Adenocarcinoma Cells) |
460 |
0.30 |
Functional ≤ 10μM
|
|
Z80362 |
Z80362
|
P388 (Lymphoma Cells) |
12 |
0.37 |
Functional ≤ 10μM
|
|
Z101973 |
Z101973
|
Paracentrotus Lividus |
20 |
0.36 |
Functional ≤ 10μM
|
|
Z80390 |
Z80390
|
PC-3 (Prostate Carcinoma Cells) |
10 |
0.37 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
10000 |
0.23 |
Functional ≤ 10μM
|
|
Z50426 |
Z50426
|
Plasmodium Falciparum (isolate K1 / Thailand) |
12 |
0.37 |
Functional ≤ 10μM
|
|
Z80414 |
Z80414
|
Raji (B-lymphoblastic Cells) |
18 |
0.36 |
Functional ≤ 10μM
|
|
Z80427 |
Z80427
|
RKO (Colon Carcinoma) |
29 |
0.35 |
Functional ≤ 10μM
|
|
Q862F3_BOVIN |
Q862F3
|
Tubulin Alpha Chain, Bovin |
1200 |
0.28 |
Functional ≤ 10μM
|
|
TBA1A_RAT |
P68370
|
Tubulin Alpha-1 Chain, Rat |
350 |
0.30 |
Functional ≤ 10μM
|
|
Q862L2_BOVIN |
Q862L2
|
Tubulin Alpha-1 Chain, Bovin |
1200 |
0.28 |
Functional ≤ 10μM
|
|
TBB2B_BOVIN |
Q6B856
|
Tubulin Beta Chain, Bovin |
1200 |
0.28 |
Functional ≤ 10μM
|
|
TBB1_HUMAN |
Q9H4B7
|
Tubulin Beta-1 Chain, Human |
350 |
0.30 |
Functional ≤ 10μM
|
|
Z50651 |
Z50651
|
Vesicular Stomatitis Virus |
100 |
0.33 |
Functional ≤ 10μM
|
|
Z100695 |
Z100695
|
WRL68 (Embryonic Hepatoma Cells) |
4800 |
0.25 |
Functional ≤ 10μM
|
|
Z80052 |
Z80052
|
C8166 (Leukemic T-cells) |
150 |
0.32 |
ADME/T ≤ 10μM
|
|
Z81115 |
Z81115
|
KB (Squamous Cell Carcinoma) |
3 |
0.40 |
ADME/T ≤ 10μM
|
|
Z81135 |
Z81135
|
L6 (Skeletal Muscle Myoblast Cells) |
10 |
0.37 |
ADME/T ≤ 10μM
|
|
Z81170 |
Z81170
|
LNCaP (Prostate Carcinoma) |
31 |
0.35 |
ADME/T ≤ 10μM
|
|
Z80390 |
Z80390
|
PC-3 (Prostate Carcinoma Cells) |
12.5 |
0.37 |
ADME/T ≤ 10μM
|
|
Z80583 |
Z80583
|
Vero (Kidney Cells) |
16 |
0.36 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.32 |
4.95 |
-16.81 |
1 |
8 |
0 |
93 |
414.41 |
4 |
↓
|
|
|
|
Analogs
-
896755
-
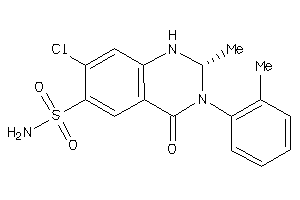
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.77 |
3.79 |
-17.4 |
3 |
6 |
0 |
93 |
365.842 |
2 |
↓
|
|
|
|
Analogs
-
601254
-
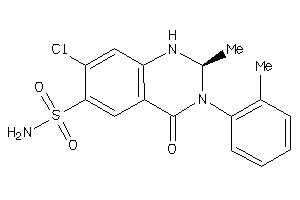
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.77 |
3.71 |
-17.08 |
3 |
6 |
0 |
93 |
365.842 |
2 |
↓
|
|
|
|
Analogs
-
1639
-
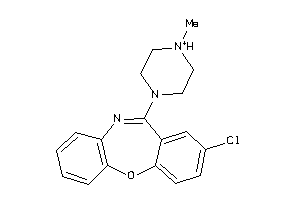
-
931
-
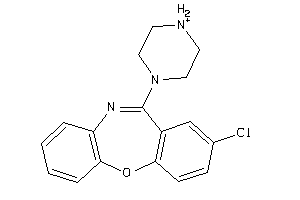
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.50 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1000 |
0.37 |
Binding ≤ 10μM
|
|
5HT6R-2-E |
Serotonin 6 (5-HT6) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
50 |
0.44 |
Binding ≤ 10μM
|
|
5HT7R-2-E |
Serotonin 7 (5-HT7) Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
43 |
0.45 |
Binding ≤ 10μM
|
|
ACM1-1-E |
Muscarinic Acetylcholine Receptor M1 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5500 |
0.32 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
22 |
0.47 |
Binding ≤ 10μM
|
|
DRD4-2-E |
Dopamine D4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.51 |
Binding ≤ 10μM
|
|
HRH1-1-E |
Histamine H1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.37 |
Binding ≤ 10μM
|
|
DRD2-15-E |
Dopamine D2 Receptor (cluster #15 Of 24), Eukaryotic |
Eukaryotes |
21 |
0.47 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
18 |
0.47 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
5012 |
0.32 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
18 |
0.47 |
Binding ≤ 1μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
21 |
0.47 |
Binding ≤ 1μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
21 |
0.47 |
Binding ≤ 1μM
|
|
DRD3_HUMAN |
P35462
|
Dopamine D3 Receptor, Human |
22 |
0.47 |
Binding ≤ 1μM
|
|
DRD4_HUMAN |
P21917
|
Dopamine D4 Receptor, Human |
14 |
0.48 |
Binding ≤ 1μM
|
|
HRH1_RAT |
P31390
|
Histamine H1 Receptor, Rat |
1000 |
0.37 |
Binding ≤ 1μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
6 |
0.50 |
Binding ≤ 1μM
|
|
5HT2C_RAT |
P08909
|
Serotonin 2c (5-HT2c) Receptor, Rat |
1000 |
0.37 |
Binding ≤ 1μM
|
|
5HT6R_HUMAN |
P50406
|
Serotonin 6 (5-HT6) Receptor, Human |
15 |
0.48 |
Binding ≤ 1μM
|
|
5HT7R_RAT |
P32305
|
Serotonin 7 (5-HT7) Receptor, Rat |
43 |
0.45 |
Binding ≤ 1μM
|
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
18 |
0.47 |
Binding ≤ 10μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
21 |
0.47 |
Binding ≤ 10μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
21 |
0.47 |
Binding ≤ 10μM
|
|
DRD3_HUMAN |
P35462
|
Dopamine D3 Receptor, Human |
22 |
0.47 |
Binding ≤ 10μM
|
|
DRD4_HUMAN |
P21917
|
Dopamine D4 Receptor, Human |
14 |
0.48 |
Binding ≤ 10μM
|
|
HRH1_RAT |
P31390
|
Histamine H1 Receptor, Rat |
1000 |
0.37 |
Binding ≤ 10μM
|
|
ACM1_HUMAN |
P11229
|
Muscarinic Acetylcholine Receptor M1, Human |
5500 |
0.32 |
Binding ≤ 10μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
6 |
0.50 |
Binding ≤ 10μM
|
|
5HT2C_RAT |
P08909
|
Serotonin 2c (5-HT2c) Receptor, Rat |
1000 |
0.37 |
Binding ≤ 10μM
|
|
5HT6R_HUMAN |
P50406
|
Serotonin 6 (5-HT6) Receptor, Human |
15 |
0.48 |
Binding ≤ 10μM
|
|
5HT7R_RAT |
P32305
|
Serotonin 7 (5-HT7) Receptor, Rat |
43 |
0.45 |
Binding ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
5011.87234 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.25 |
7.92 |
-8.16 |
0 |
4 |
0 |
33 |
327.815 |
1 |
↓
|
|
|
|
Analogs
-
22001248
-
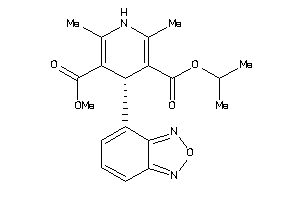
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
8.59 |
-22.3 |
1 |
8 |
0 |
104 |
371.393 |
6 |
↓
|
|
|
|
Analogs
-
607939
-
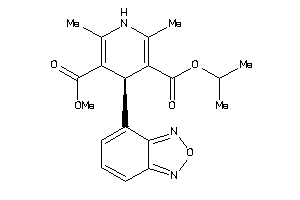
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
8.58 |
-22.86 |
1 |
8 |
0 |
104 |
371.393 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-10-E |
Acetylcholinesterase (cluster #10 Of 12), Eukaryotic |
Eukaryotes |
5360 |
0.61 |
Binding ≤ 10μM
|
|
Z50512-3-O |
Cavia Porcellus (cluster #3 Of 7), Other |
Other |
2770 |
0.65 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.06 |
4.96 |
-29.63 |
1 |
2 |
1 |
20 |
166.244 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.06 |
5.72 |
-41.43 |
0 |
2 |
0 |
23 |
165.236 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.44 |
3.11 |
-13.33 |
0 |
4 |
0 |
54 |
273.741 |
1 |
↓
|
|
|
|
Analogs
-
2944385
-
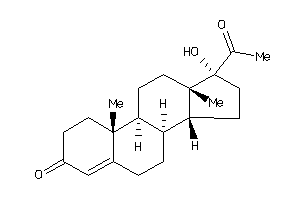
-
3651055
-
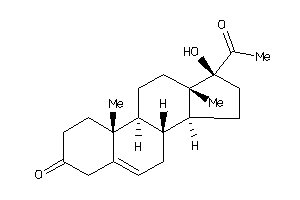
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.86 |
7.53 |
-9.7 |
1 |
3 |
0 |
54 |
330.468 |
1 |
↓
|
|
|
|
Analogs
-
2944385
-

-
3651055
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.86 |
8.23 |
-10.11 |
1 |
3 |
0 |
54 |
330.468 |
1 |
↓
|
|
|
|
Analogs
-
2944385
-

-
3651055
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.86 |
7.36 |
-9.61 |
1 |
3 |
0 |
54 |
330.468 |
1 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
496649
-
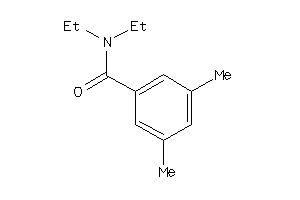
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.08 |
6.93 |
-8.5 |
0 |
2 |
0 |
20 |
191.274 |
3 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.11 |
10.68 |
-12.93 |
1 |
5 |
0 |
73 |
390.52 |
7 |
↓
|
|
|
|
|
|
|
Analogs
-
3831450
-
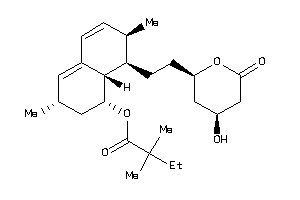
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HMDH-1-E |
HMG-CoA Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
28 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.11 |
10.52 |
-8.63 |
1 |
5 |
0 |
73 |
390.52 |
7 |
↓
|
|
|
|
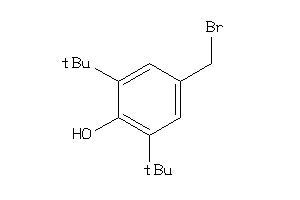
Analogs
-
2577829
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.43 |
7.87 |
-2.46 |
1 |
1 |
0 |
20 |
220.356 |
2 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
5.14 |
-8.02 |
1 |
3 |
0 |
47 |
242.274 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.81 |
5.92 |
-47.76 |
0 |
3 |
-1 |
49 |
241.266 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.31 |
5.39 |
-6.68 |
2 |
3 |
0 |
52 |
193.246 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.49 |
2.08 |
-8.39 |
0 |
4 |
0 |
42 |
192.218 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.18 |
3.38 |
-2.48 |
0 |
6 |
0 |
66 |
232.41 |
0 |
↓
|
|
|
|
|
|
|
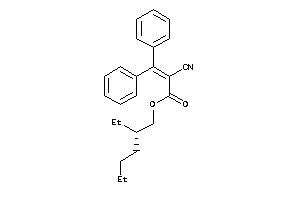
Analogs
-
4975824
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.54 |
15.46 |
-5.79 |
0 |
3 |
0 |
50 |
361.485 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.08 |
-7.53 |
-18.4 |
7 |
10 |
0 |
177 |
434.397 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.08 |
-6.54 |
-69.77 |
6 |
10 |
-1 |
180 |
433.389 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.08 |
-6.54 |
-65.63 |
6 |
10 |
-1 |
180 |
433.389 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.70 |
0.35 |
-6.78 |
2 |
2 |
0 |
43 |
101.149 |
2 |
↓
|
|
|
|
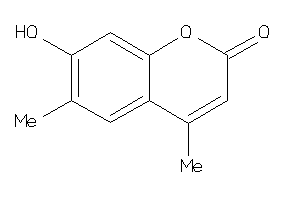
Analogs
-
34480190
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DHB3-2-E |
Estradiol 17-beta-dehydrogenase 3 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1000 |
0.65 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.89 |
3.72 |
-12.48 |
1 |
3 |
0 |
50 |
176.171 |
0 |
↓
|
|
|
|
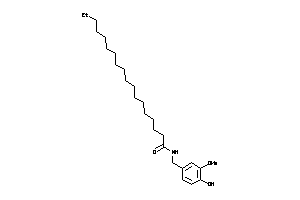
Analogs
-
43273992
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.12 |
-1.49 |
-9.46 |
2 |
4 |
0 |
58 |
293.407 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.01 |
7.11 |
-6.01 |
1 |
1 |
0 |
16 |
199.278 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.86 |
2.94 |
-8.03 |
1 |
2 |
0 |
37 |
150.177 |
2 |
↓
|
|
|
|
Analogs
-
33804374
-
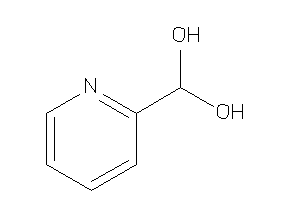
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.11 |
0.18 |
-5.47 |
1 |
2 |
0 |
33 |
109.128 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.11 |
0.59 |
-30.28 |
2 |
2 |
1 |
34 |
110.136 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
2940 |
0.86 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
2940 |
0.86 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.30 |
1.63 |
-5.69 |
1 |
2 |
0 |
29 |
124.139 |
1 |
↓
|
|
|
|
Analogs
-
12409058
-
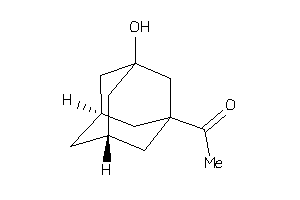
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.14 |
2.39 |
-5.27 |
1 |
2 |
0 |
37 |
166.22 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.53 |
2.75 |
-4.47 |
0 |
1 |
0 |
9 |
126.077 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.49 |
-0.03 |
-3.97 |
2 |
2 |
0 |
40 |
118.176 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.49 |
-0.39 |
-4.1 |
2 |
2 |
0 |
40 |
118.176 |
2 |
↓
|
|
|
|
Analogs
-
39343114
-
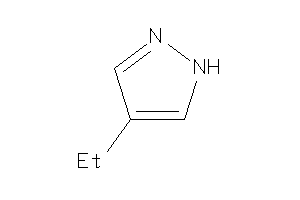
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.70 |
1.64 |
-4.93 |
1 |
2 |
0 |
29 |
82.106 |
0 |
↓
|
|
|
|
Analogs
-
1238584
-
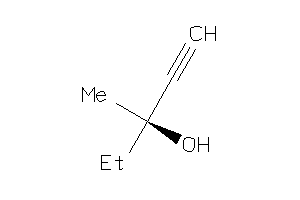
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.15 |
1.44 |
-3.1 |
1 |
1 |
0 |
20 |
98.145 |
1 |
↓
|
|
|
|
Analogs
-
1238583
-
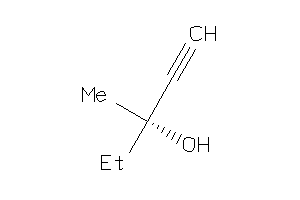
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.15 |
1.4 |
-3.15 |
1 |
1 |
0 |
20 |
98.145 |
1 |
↓
|
|