|
|
|
|
|
|
|
|
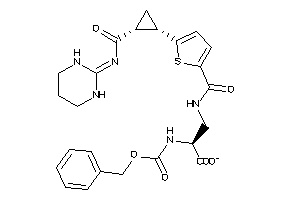
Analogs
-
26145951
-
-
26145936
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ITA2B-1-E |
Integrin Alpha-IIb (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2450 |
0.22 |
Binding ≤ 10μM
|
|
ITB3-1-E |
Integrin Beta-3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2450 |
0.22 |
Binding ≤ 10μM
|
|
Z104292-1-O |
Integrin Alpha-V/beta-3 (cluster #1 Of 1), Other |
Other |
60 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.59 |
8.01 |
-56.96 |
4 |
11 |
-1 |
161 |
512.568 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.59 |
7.85 |
-80.64 |
5 |
11 |
0 |
163 |
513.576 |
11 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.59 |
9.08 |
-73.25 |
5 |
11 |
0 |
163 |
513.576 |
10 |
↓
|
|
|
|
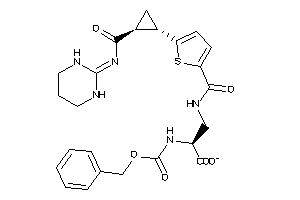
Analogs
-
26145943
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ITA2B-1-E |
Integrin Alpha-IIb (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2450 |
0.22 |
Binding ≤ 10μM
|
|
ITB3-1-E |
Integrin Beta-3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2450 |
0.22 |
Binding ≤ 10μM
|
|
Z104292-1-O |
Integrin Alpha-V/beta-3 (cluster #1 Of 1), Other |
Other |
60 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.59 |
8.03 |
-58.02 |
4 |
11 |
-1 |
161 |
512.568 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.59 |
7.85 |
-79.2 |
5 |
11 |
0 |
163 |
513.576 |
11 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.59 |
9.1 |
-72.13 |
5 |
11 |
0 |
163 |
513.576 |
10 |
↓
|
|
|
|
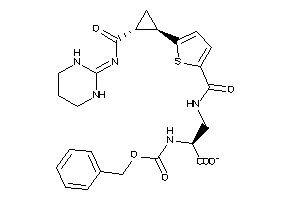
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.85 |
21.27 |
-58.95 |
2 |
9 |
0 |
114 |
643.91 |
18 |
↓
|
|
Hi
High (pH 8-9.5)
|
8.85 |
20.39 |
-48.32 |
1 |
9 |
-1 |
113 |
642.902 |
18 |
↓
|
|
Lo
Low (pH 4.5-6)
|
8.85 |
19.29 |
-26.74 |
3 |
9 |
1 |
111 |
644.918 |
18 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
9.05 |
22.05 |
-59.2 |
2 |
9 |
0 |
114 |
657.937 |
19 |
↓
|
|
Hi
High (pH 8-9.5)
|
9.05 |
21.17 |
-48.3 |
1 |
9 |
-1 |
113 |
656.929 |
19 |
↓
|
|
Lo
Low (pH 4.5-6)
|
9.05 |
20.07 |
-26.74 |
3 |
9 |
1 |
111 |
658.945 |
19 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
9.05 |
22.12 |
-61.21 |
2 |
9 |
0 |
114 |
657.937 |
19 |
↓
|
|
Hi
High (pH 8-9.5)
|
9.05 |
20.71 |
-51.99 |
1 |
9 |
-1 |
113 |
656.929 |
19 |
↓
|
|
Lo
Low (pH 4.5-6)
|
9.05 |
20.14 |
-28.05 |
3 |
9 |
1 |
111 |
658.945 |
19 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CNR1-1-E |
Cannabinoid CB1 Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
624 |
0.33 |
Binding ≤ 10μM
|
|
CNR2-1-E |
Cannabinoid CB2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
155 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.14 |
12.94 |
-8.97 |
1 |
4 |
0 |
45 |
367.518 |
2 |
↓
|
|