|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9500 |
0.17 |
Binding ≤ 10μM
|
|
PLMN-2-E |
Plasminogen (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.18 |
Binding ≤ 10μM
|
|
ST14-1-E |
Matriptase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.30 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
6000 |
0.18 |
Binding ≤ 10μM
|
|
UROK-2-E |
Urokinase-type Plasminogen Activator (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1700 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.17 |
1.97 |
-169.14 |
14 |
13 |
3 |
238 |
588.759 |
14 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PLMN-2-E |
Plasminogen (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.19 |
Binding ≤ 10μM
|
|
ST14-1-E |
Matriptase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
255 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.01 |
4.92 |
-157.99 |
9 |
12 |
1 |
215 |
587.723 |
14 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.01 |
2.94 |
-130.68 |
10 |
12 |
2 |
212 |
588.731 |
14 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6100 |
0.17 |
Binding ≤ 10μM
|
|
PLMN-2-E |
Plasminogen (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2800 |
0.19 |
Binding ≤ 10μM
|
|
ST14-1-E |
Matriptase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.27 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
600 |
0.21 |
Binding ≤ 10μM
|
|
UROK-2-E |
Urokinase-type Plasminogen Activator (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2500 |
0.19 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.30 |
4.94 |
-136.31 |
9 |
12 |
2 |
201 |
602.758 |
15 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.17 |
Binding ≤ 10μM
|
|
PLMN-2-E |
Plasminogen (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.19 |
Binding ≤ 10μM
|
|
ST14-1-E |
Matriptase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.27 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
850 |
0.20 |
Binding ≤ 10μM
|
|
UROK-2-E |
Urokinase-type Plasminogen Activator (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3300 |
0.18 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.15 |
2.47 |
-138.94 |
10 |
12 |
2 |
204 |
601.774 |
14 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8200 |
0.17 |
Binding ≤ 10μM
|
|
PLMN-2-E |
Plasminogen (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2900 |
0.19 |
Binding ≤ 10μM
|
|
ST14-1-E |
Matriptase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
12 |
0.27 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
670 |
0.21 |
Binding ≤ 10μM
|
|
UROK-2-E |
Urokinase-type Plasminogen Activator (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2100 |
0.19 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.53 |
1.06 |
-132.77 |
11 |
12 |
2 |
218 |
587.747 |
14 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.21 |
Binding ≤ 10μM
|
|
PLMN-2-E |
Plasminogen (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1200 |
0.22 |
Binding ≤ 10μM
|
|
ST14-1-E |
Matriptase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
13 |
0.29 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1250 |
0.22 |
Binding ≤ 10μM
|
|
UROK-2-E |
Urokinase-type Plasminogen Activator (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3300 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.76 |
-1.14 |
-130.35 |
11 |
12 |
2 |
218 |
545.666 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PLMN-2-E |
Plasminogen (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1300 |
0.22 |
Binding ≤ 10μM
|
|
ST14-1-E |
Matriptase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
31 |
0.28 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
2100 |
0.21 |
Binding ≤ 10μM
|
|
UROK-2-E |
Urokinase-type Plasminogen Activator (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
7400 |
0.19 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.76 |
-1.02 |
-137.68 |
11 |
12 |
2 |
218 |
545.666 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PLMN-2-E |
Plasminogen (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1300 |
0.22 |
Binding ≤ 10μM
|
|
ST14-1-E |
Matriptase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
31 |
0.28 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
2100 |
0.21 |
Binding ≤ 10μM
|
|
UROK-2-E |
Urokinase-type Plasminogen Activator (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
7400 |
0.19 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.76 |
-1.02 |
-141.17 |
11 |
12 |
2 |
218 |
545.666 |
11 |
↓
|
|
|
|
Analogs
-
40955895
-
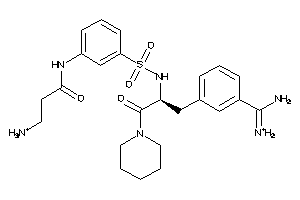
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.20 |
Binding ≤ 10μM
|
|
PLMN-2-E |
Plasminogen (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3300 |
0.22 |
Binding ≤ 10μM
|
|
ST14-1-E |
Matriptase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
33 |
0.30 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
270 |
0.26 |
Binding ≤ 10μM
|
|
UROK-2-E |
Urokinase-type Plasminogen Activator (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2600 |
0.22 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.35 |
2.35 |
-131.3 |
9 |
10 |
2 |
175 |
502.641 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4400 |
0.21 |
Binding ≤ 10μM
|
|
PLMN-2-E |
Plasminogen (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6200 |
0.20 |
Binding ≤ 10μM
|
|
ST14-1-E |
Matriptase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
53 |
0.28 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
110 |
0.27 |
Binding ≤ 10μM
|
|
UROK-2-E |
Urokinase-type Plasminogen Activator (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
3000 |
0.21 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.11 |
2.72 |
-130.49 |
9 |
10 |
2 |
175 |
516.668 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PLMN-2-E |
Plasminogen (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9400 |
0.19 |
Binding ≤ 10μM
|
|
ST14-1-E |
Matriptase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
180 |
0.25 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
2300 |
0.21 |
Binding ≤ 10μM
|
|
UROK-2-E |
Urokinase-type Plasminogen Activator (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
7300 |
0.19 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.58 |
3.76 |
-172.35 |
9 |
12 |
1 |
215 |
545.642 |
11 |
↓
|
|
|
|
Analogs
-
40952844
-
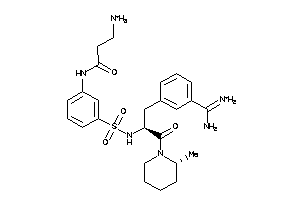
-
40952845
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PLMN-2-E |
Plasminogen (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4500 |
0.21 |
Binding ≤ 10μM
|
|
ST14-1-E |
Matriptase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
470 |
0.25 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
395 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.02 |
2.74 |
-129.61 |
9 |
10 |
2 |
175 |
516.668 |
10 |
↓
|
|
|
|
Analogs
-
40952844
-

-
40952845
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PLMN-2-E |
Plasminogen (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4500 |
0.21 |
Binding ≤ 10μM
|
|
ST14-1-E |
Matriptase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
470 |
0.25 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
395 |
0.25 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.02 |
2.74 |
-128.96 |
9 |
10 |
2 |
175 |
516.668 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ST14-1-E |
Matriptase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2400 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.50 |
3.71 |
-137.39 |
9 |
12 |
2 |
201 |
560.677 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.41 |
7.92 |
-17.5 |
1 |
5 |
0 |
66 |
404.507 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.41 |
7.51 |
-14.85 |
1 |
5 |
0 |
66 |
404.507 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.41 |
7.89 |
-14.03 |
1 |
5 |
0 |
66 |
404.507 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.41 |
7.9 |
-13.68 |
1 |
5 |
0 |
66 |
404.507 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.89 |
8.1 |
-14.59 |
1 |
5 |
0 |
66 |
418.534 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.89 |
8.33 |
-14.89 |
1 |
5 |
0 |
66 |
418.534 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.89 |
8.34 |
-14.5 |
1 |
5 |
0 |
66 |
418.534 |
6 |
↓
|
|