|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.96 |
6.26 |
-46.47 |
1 |
5 |
-1 |
78 |
309.169 |
4 |
↓
|
|
|
|
Analogs
-
38613343
-
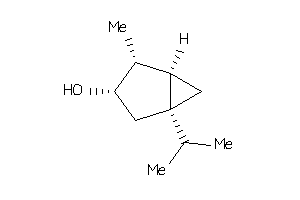
-
12153566
-
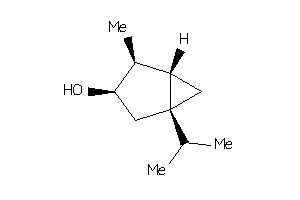
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.88 |
5.97 |
-2.29 |
1 |
1 |
0 |
20 |
222.372 |
1 |
↓
|
|
|
|
Analogs
-
38613343
-

-
12153566
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.88 |
5.88 |
-2.25 |
1 |
1 |
0 |
20 |
222.372 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.36 |
6.87 |
-3.03 |
1 |
1 |
0 |
20 |
216.324 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.36 |
6.79 |
-3.24 |
1 |
1 |
0 |
20 |
216.324 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.10 |
7.15 |
-2.87 |
1 |
1 |
0 |
20 |
295.22 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.66 |
11.12 |
-6.18 |
0 |
2 |
0 |
26 |
337.257 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.89 |
7.65 |
-2.88 |
1 |
1 |
0 |
20 |
295.22 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.70 |
3.21 |
-106.25 |
4 |
7 |
-1 |
137 |
297.331 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.55 |
4.88 |
-107.46 |
4 |
7 |
-1 |
137 |
331.348 |
6 |
↓
|
|
|
|
Analogs
-
3979742
-
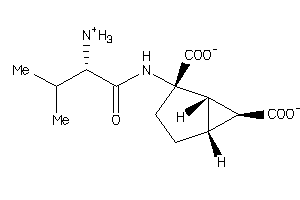
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.07 |
1.73 |
-111.54 |
4 |
7 |
-1 |
137 |
255.25 |
4 |
↓
|
|
|
|
Analogs
-
3979743
-
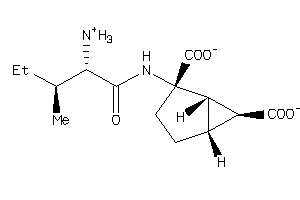
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.76 |
3.72 |
-108.4 |
4 |
7 |
-1 |
137 |
297.331 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.80 |
5.2 |
-113.23 |
5 |
9 |
-1 |
166 |
368.41 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.80 |
4.87 |
-110.88 |
4 |
9 |
-2 |
164 |
367.402 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
34 |
0.39 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6420 |
0.27 |
Binding ≤ 10μM
|
|
AA3R-2-E |
Adenosine Receptor A3 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
13 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.42 |
3.19 |
-15.92 |
4 |
9 |
0 |
125 |
372.429 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R_HUMAN |
P30542
|
Adenosine A1 Receptor, Human |
18.3 |
0.39 |
Binding ≤ 1μM
|
|
AA1R_RAT |
P25099
|
Adenosine A1 Receptor, Rat |
17.4 |
0.39 |
Binding ≤ 1μM
|
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
3.7 |
0.42 |
Binding ≤ 1μM
|
|
AA3R_RAT |
P28647
|
Adenosine A3 Receptor, Rat |
5.8 |
0.41 |
Binding ≤ 1μM
|
|
AA1R_RAT |
P25099
|
Adenosine A1 Receptor, Rat |
17.4 |
0.39 |
Binding ≤ 10μM
|
|
AA1R_HUMAN |
P30542
|
Adenosine A1 Receptor, Human |
18.3 |
0.39 |
Binding ≤ 10μM
|
|
AA2AR_HUMAN |
P29274
|
Adenosine A2a Receptor, Human |
3250 |
0.27 |
Binding ≤ 10μM
|
|
AA3R_RAT |
P28647
|
Adenosine A3 Receptor, Rat |
5.8 |
0.41 |
Binding ≤ 10μM
|
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
3.7 |
0.42 |
Binding ≤ 10μM
|
|
AA1R_HUMAN |
P30542
|
Adenosine A1 Receptor, Human |
8.2 |
0.40 |
Functional ≤ 10μM
|
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
2.8 |
0.43 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.42 |
2.99 |
-15.24 |
4 |
9 |
0 |
125 |
406.874 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
190 |
0.31 |
Binding ≤ 10μM
|
|
AA3R-2-E |
Adenosine Receptor A3 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
15 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.56 |
4.21 |
-15.07 |
4 |
9 |
0 |
125 |
432.912 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1S,2R,3S,4R,5R)-2-(2,4-dioxopyrimidin-1-yl)-3,4-dihydroxy-5-bicyclo[3.1.0]hexanyl]methyl
[(1S,2R,3S,4R,5R)-2-(2,4-dioxopy…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.00 |
-2.73 |
-256.47 |
3 |
13 |
-3 |
217 |
411.176 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.00 |
-3.89 |
-135.92 |
4 |
13 |
-2 |
214 |
412.184 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RY6-1-E |
Pyrimidinergic Receptor P2Y6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
230 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.71 |
-0.44 |
-242.35 |
2 |
12 |
-3 |
197 |
395.177 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.71 |
-1.6 |
-124.48 |
3 |
12 |
-2 |
194 |
396.185 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.36 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
631 |
0.26 |
Binding ≤ 10μM
|
|
DRD2-2-E |
Dopamine D2 Receptor (cluster #2 Of 24), Eukaryotic |
Eukaryotes |
501 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.75 |
11.7 |
-58.92 |
2 |
6 |
1 |
73 |
478.564 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
251 |
0.28 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2512 |
0.24 |
Binding ≤ 10μM
|
|
DRD2-2-E |
Dopamine D2 Receptor (cluster #2 Of 24), Eukaryotic |
Eukaryotes |
3162 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.75 |
11.72 |
-59.27 |
2 |
6 |
1 |
73 |
478.564 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.35 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
631 |
0.26 |
Binding ≤ 10μM
|
|
DRD2-2-E |
Dopamine D2 Receptor (cluster #2 Of 24), Eukaryotic |
Eukaryotes |
200 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.84 |
11.67 |
-60.78 |
2 |
6 |
1 |
73 |
496.554 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
32 |
0.31 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1000 |
0.25 |
Binding ≤ 10μM
|
|
DRD2-2-E |
Dopamine D2 Receptor (cluster #2 Of 24), Eukaryotic |
Eukaryotes |
3162 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.84 |
11.74 |
-57.1 |
2 |
6 |
1 |
73 |
496.554 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MCHR1-1-E |
Melanin-concentrating Hormone Receptor 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.32 |
Binding ≤ 10μM
|
|
CP2CJ-1-E |
Cytochrome P450 2C19 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6100 |
0.19 |
ADME/T ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
50 |
0.26 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.29 |
14.34 |
-49.69 |
2 |
6 |
1 |
64 |
544.617 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.29 |
11.97 |
-12.21 |
1 |
6 |
0 |
63 |
543.609 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.29 |
14.24 |
-59.45 |
2 |
6 |
1 |
64 |
544.617 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1R,2R,3R,4S,5S)-2-(2,4-dioxopyrimidin-1-yl)-3,4-dihydroxy-5-bicyclo[3.1.0]hexanyl]methyl
[(1R,2R,3R,4S,5S)-2-(2,4-dioxopy…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.00 |
-1.66 |
-242.88 |
3 |
13 |
-3 |
217 |
411.176 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.00 |
-2.81 |
-126.69 |
4 |
13 |
-2 |
214 |
412.184 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1R,2S,3R,4S,5S)-2-(2,4-dioxopyrimidin-1-yl)-3,4-dihydroxy-5-bicyclo[3.1.0]hexanyl]methyl
[(1R,2S,3R,4S,5S)-2-(2,4-dioxopy…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.00 |
-2.76 |
-255.74 |
3 |
13 |
-3 |
217 |
411.176 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.00 |
-3.92 |
-135.43 |
4 |
13 |
-2 |
214 |
412.184 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.20 |
1.05 |
-10.9 |
4 |
7 |
0 |
110 |
261.285 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.20 |
0.73 |
-10.89 |
4 |
7 |
0 |
110 |
261.285 |
2 |
↓
|
|