|
|
Analogs
-
34633666
-
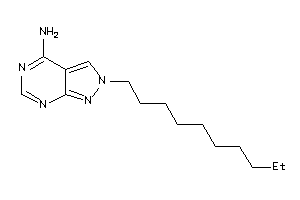
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.17 |
11.16 |
-16.42 |
2 |
5 |
0 |
70 |
303.454 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SOAT1-1-E |
Acyl Coenzyme A:cholesterol Acyltransferase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4350 |
0.26 |
Binding ≤ 10μM |
|
SOAT2-1-E |
Acyl Coenzyme A:cholesterol Acyltransferase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7130 |
0.25 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.65 |
12.65 |
-19.47 |
2 |
7 |
0 |
87 |
420.627 |
14 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SOAT1-1-E |
Acyl Coenzyme A:cholesterol Acyltransferase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4230 |
0.21 |
Binding ≤ 10μM |
|
SOAT2-1-E |
Acyl Coenzyme A:cholesterol Acyltransferase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4230 |
0.21 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.73 |
13.11 |
-22.22 |
4 |
8 |
0 |
113 |
505.639 |
14 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SOAT1-1-E |
Acyl Coenzyme A:cholesterol Acyltransferase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3310 |
0.23 |
Binding ≤ 10μM |
|
SOAT2-1-E |
Acyl Coenzyme A:cholesterol Acyltransferase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
710 |
0.25 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.37 |
14.87 |
-17.98 |
2 |
7 |
0 |
87 |
490.624 |
14 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.29 |
6.38 |
-17.95 |
2 |
5 |
0 |
70 |
225.255 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.29 |
2.29 |
-40.37 |
3 |
5 |
1 |
71 |
226.263 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.50 |
7.15 |
-16.52 |
2 |
5 |
0 |
70 |
239.282 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.50 |
3.06 |
-39.21 |
3 |
5 |
1 |
71 |
240.29 |
3 |
↓
|
|
|
|
Analogs
-
34633667
-
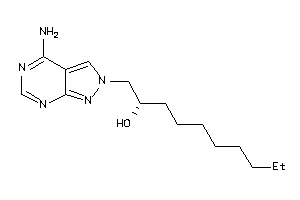
-
34633668
-
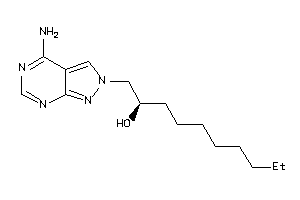
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA-1-E |
Adenosine Deaminase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.66 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.00 |
4.76 |
-20.19 |
3 |
6 |
0 |
90 |
263.345 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.00 |
0.66 |
-41.15 |
4 |
6 |
1 |
91 |
264.353 |
7 |
↓
|
|
|
|
Analogs
-
34633667
-

-
34633668
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA-1-E |
Adenosine Deaminase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.66 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.00 |
4.97 |
-19.2 |
3 |
6 |
0 |
90 |
263.345 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.00 |
5.23 |
-34.09 |
4 |
6 |
1 |
91 |
264.353 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.00 |
0.87 |
-40.3 |
4 |
6 |
1 |
91 |
264.353 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA-1-E |
Adenosine Deaminase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
53 |
0.51 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
53 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.37 |
5.5 |
-17.19 |
3 |
6 |
0 |
90 |
277.372 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.37 |
5.77 |
-34 |
4 |
6 |
1 |
91 |
278.38 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.37 |
1.41 |
-40.06 |
4 |
6 |
1 |
91 |
278.38 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA-1-E |
Adenosine Deaminase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
53 |
0.51 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
53 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.37 |
5.39 |
-16.48 |
3 |
6 |
0 |
90 |
277.372 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.37 |
1.3 |
-39.12 |
4 |
6 |
1 |
91 |
278.38 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA-1-E |
Adenosine Deaminase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
53 |
0.51 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
53 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.37 |
5.3 |
-14.57 |
3 |
6 |
0 |
90 |
277.372 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.37 |
5.57 |
-29.51 |
4 |
6 |
1 |
91 |
278.38 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.37 |
1.21 |
-34.53 |
4 |
6 |
1 |
91 |
278.38 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA-1-E |
Adenosine Deaminase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
53 |
0.51 |
Binding ≤ 10μM
|
|
VGFR2-1-E |
Vascular Endothelial Growth Factor Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
53 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.37 |
5.43 |
-17.11 |
3 |
6 |
0 |
90 |
277.372 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.37 |
5.7 |
-33.61 |
4 |
6 |
1 |
91 |
278.38 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.37 |
1.34 |
-39.48 |
4 |
6 |
1 |
91 |
278.38 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.29 |
Binding ≤ 10μM
|
|
BCR-1-E |
Breakpoint Cluster Region Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.53 |
9.57 |
-19.05 |
1 |
7 |
0 |
78 |
386.337 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1320 |
0.34 |
Binding ≤ 10μM
|
|
BCR-1-E |
Breakpoint Cluster Region Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1320 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.54 |
5.01 |
-16.06 |
2 |
7 |
0 |
85 |
339.277 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABL1-1-E |
Tyrosine-protein Kinase ABL (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.36 |
Binding ≤ 10μM
|
|
BCR-1-E |
Breakpoint Cluster Region Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1200 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.55 |
8.35 |
-16.33 |
1 |
6 |
0 |
65 |
323.278 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.24 |
6.09 |
-22.57 |
1 |
8 |
0 |
89 |
337.387 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.24 |
6.5 |
-43.36 |
2 |
8 |
1 |
90 |
338.395 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.41 |
3.7 |
-42.52 |
1 |
6 |
-1 |
87 |
333.418 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.95 |
5.83 |
-14.58 |
2 |
6 |
0 |
84 |
334.426 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.95 |
5.63 |
-12.29 |
2 |
6 |
0 |
84 |
334.426 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.79 |
4.36 |
-47.45 |
1 |
6 |
-1 |
87 |
315.378 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.33 |
6.32 |
-12.49 |
2 |
6 |
0 |
84 |
316.386 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.95 |
2.76 |
-5.2 |
1 |
4 |
0 |
54 |
199.011 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
312 |
0.40 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
121 |
0.42 |
Binding ≤ 10μM
|
|
AA3R-2-E |
Adenosine Receptor A3 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
93 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.40 |
9.72 |
-20.18 |
2 |
5 |
0 |
70 |
301.353 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.75 |
10.26 |
-25.6 |
1 |
7 |
0 |
82 |
359.389 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.47 |
11.58 |
-31.66 |
3 |
7 |
1 |
92 |
446 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.94 |
11.5 |
-14.53 |
2 |
7 |
0 |
91 |
444.992 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.47 |
11.66 |
-33.64 |
3 |
7 |
1 |
92 |
446 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.94 |
11.6 |
-16.18 |
2 |
7 |
0 |
91 |
444.992 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.26 |
8.95 |
-31.97 |
4 |
7 |
1 |
101 |
385.517 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.00 |
8.9 |
-12.79 |
3 |
7 |
0 |
99 |
384.509 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.49 |
9.19 |
-31.67 |
4 |
7 |
1 |
101 |
385.517 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.77 |
9.14 |
-12.81 |
3 |
7 |
0 |
99 |
384.509 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.49 |
9.18 |
-31.71 |
4 |
7 |
1 |
101 |
385.517 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.77 |
9.14 |
-12.81 |
3 |
7 |
0 |
99 |
384.509 |
6 |
↓
|
|
|
|
Analogs
-
33686123
-
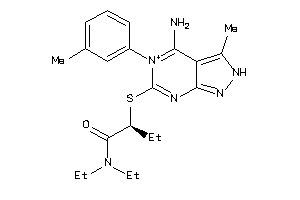
-
33686124
-
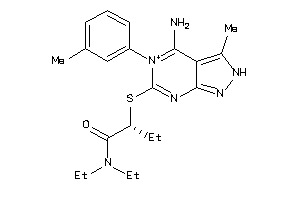
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.60 |
10.37 |
-31.56 |
4 |
7 |
1 |
101 |
413.571 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.87 |
10.3 |
-15.37 |
3 |
7 |
0 |
99 |
412.563 |
8 |
↓
|
|
|
|
Analogs
-
33686123
-

-
33686124
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.19 |
11.14 |
-31.9 |
3 |
7 |
1 |
92 |
411.555 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.08 |
11.08 |
-16.57 |
2 |
7 |
0 |
91 |
410.547 |
5 |
↓
|
|
|
|
Analogs
-
33672629
-
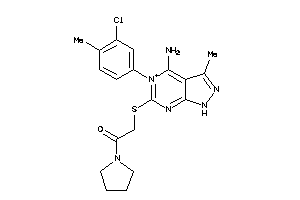
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.39 |
9.32 |
-35.04 |
4 |
7 |
1 |
101 |
419.962 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.66 |
9.26 |
-12.49 |
3 |
7 |
0 |
99 |
418.954 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.05 |
10.86 |
-34.7 |
3 |
7 |
1 |
92 |
419.962 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.22 |
10.79 |
-12.47 |
2 |
7 |
0 |
91 |
418.954 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.16 |
9.55 |
-34.8 |
4 |
7 |
1 |
101 |
419.962 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.43 |
9.49 |
-12.55 |
3 |
7 |
0 |
99 |
418.954 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.16 |
9.55 |
-34.81 |
4 |
7 |
1 |
101 |
419.962 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.43 |
9.49 |
-12.54 |
3 |
7 |
0 |
99 |
418.954 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.09 |
9.87 |
-35.6 |
3 |
8 |
1 |
101 |
461.999 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.18 |
9.85 |
-15.97 |
2 |
8 |
0 |
100 |
460.991 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.08 |
9.55 |
-35.85 |
4 |
8 |
1 |
110 |
435.533 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.34 |
9.53 |
-15.37 |
3 |
8 |
0 |
109 |
434.525 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.63 |
7.52 |
-34.53 |
4 |
8 |
1 |
110 |
401.516 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.64 |
7.45 |
-14.35 |
3 |
8 |
0 |
109 |
400.508 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.07 |
9.05 |
-34.18 |
3 |
8 |
1 |
101 |
401.516 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.20 |
8.99 |
-14.27 |
2 |
8 |
0 |
100 |
400.508 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.86 |
7.75 |
-34.27 |
4 |
8 |
1 |
110 |
401.516 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.41 |
7.69 |
-14.32 |
3 |
8 |
0 |
109 |
400.508 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.86 |
7.75 |
-34.26 |
4 |
8 |
1 |
110 |
401.516 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.41 |
7.69 |
-14.35 |
3 |
8 |
0 |
109 |
400.508 |
7 |
↓
|
|
|
|
Analogs
-
33705853
-
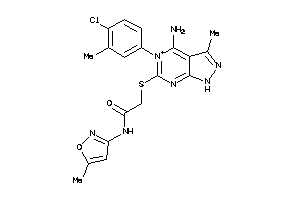
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.39 |
9.32 |
-34.44 |
4 |
7 |
1 |
101 |
419.962 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.66 |
9.25 |
-11.68 |
3 |
7 |
0 |
99 |
418.954 |
7 |
↓
|
|
|
|
Analogs
-
33705853
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.05 |
10.86 |
-34 |
3 |
7 |
1 |
92 |
419.962 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.22 |
10.79 |
-11.61 |
2 |
7 |
0 |
91 |
418.954 |
6 |
↓
|
|
|
|
Analogs
-
37866410
-
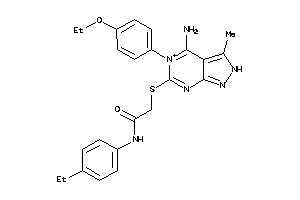
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.94 |
11.24 |
-35.11 |
4 |
8 |
1 |
110 |
463.587 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.00 |
11.25 |
-15.56 |
3 |
8 |
0 |
109 |
462.579 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.21 |
11.92 |
-32.23 |
3 |
7 |
1 |
92 |
425.582 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.26 |
11.94 |
-13.38 |
2 |
7 |
0 |
91 |
424.574 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.02 |
9.89 |
-32.31 |
4 |
7 |
1 |
101 |
399.544 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.08 |
9.9 |
-13.84 |
3 |
7 |
0 |
99 |
398.536 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.14 |
9.62 |
-33.43 |
4 |
7 |
1 |
101 |
419.962 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.40 |
9.57 |
-12.27 |
3 |
7 |
0 |
99 |
418.954 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.14 |
9.62 |
-33.39 |
4 |
7 |
1 |
101 |
419.962 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.40 |
9.57 |
-12.31 |
3 |
7 |
0 |
99 |
418.954 |
6 |
↓
|
|
|
|
Analogs
-
37867800
-
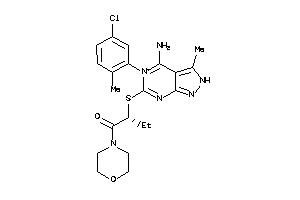
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.11 |
9.84 |
-34.29 |
3 |
8 |
1 |
101 |
461.999 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.15 |
9.76 |
-16.2 |
2 |
8 |
0 |
100 |
460.991 |
5 |
↓
|
|
|
|
Analogs
-
37867800
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.11 |
9.94 |
-32.99 |
3 |
8 |
1 |
101 |
461.999 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.15 |
9.88 |
-14.96 |
2 |
8 |
0 |
100 |
460.991 |
5 |
↓
|
|
|
|
Analogs
-
33679576
-
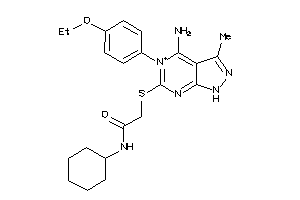
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.35 |
11.34 |
-32.31 |
3 |
8 |
1 |
101 |
455.608 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.40 |
11.32 |
-12.08 |
2 |
8 |
0 |
100 |
454.6 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.40 |
11.34 |
-13.09 |
2 |
8 |
0 |
100 |
454.6 |
7 |
↓
|
|
|
|
Analogs
-
34872893
-
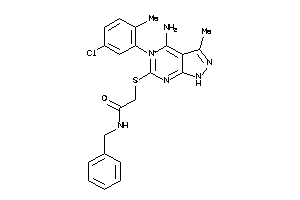
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.57 |
11.79 |
-31.8 |
4 |
7 |
1 |
101 |
447.588 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.05 |
11.73 |
-12.74 |
3 |
7 |
0 |
99 |
446.58 |
6 |
↓
|
|