|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
C11B1_HUMAN |
P15538
|
Cytochrome P450 11B1, Human |
342 |
0.45 |
Binding ≤ 1μM
|
|
C11B2_HUMAN |
P19099
|
Cytochrome P450 11B2, Human |
261 |
0.46 |
Binding ≤ 1μM
|
|
CP17A_HUMAN |
P05093
|
Cytochrome P450 17A1, Human |
97 |
0.49 |
Binding ≤ 1μM
|
|
CP19A_HUMAN |
P11511
|
Cytochrome P450 19A1, Human |
663 |
0.43 |
Binding ≤ 1μM
|
|
CP3A4_HUMAN |
P08684
|
Cytochrome P450 3A4, Human |
538 |
0.44 |
Binding ≤ 1μM
|
|
C11B1_HUMAN |
P15538
|
Cytochrome P450 11B1, Human |
342 |
0.45 |
Binding ≤ 10μM
|
|
C11B2_HUMAN |
P19099
|
Cytochrome P450 11B2, Human |
261 |
0.46 |
Binding ≤ 10μM
|
|
CP17A_HUMAN |
P05093
|
Cytochrome P450 17A1, Human |
97 |
0.49 |
Binding ≤ 10μM
|
|
CP19A_HUMAN |
P11511
|
Cytochrome P450 19A1, Human |
663 |
0.43 |
Binding ≤ 10μM
|
|
CP3A4_HUMAN |
P08684
|
Cytochrome P450 3A4, Human |
538 |
0.44 |
Binding ≤ 10μM
|
|
CP3A4_HUMAN |
P08684
|
Cytochrome P450 3A4, Human |
538 |
0.44 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.90 |
7.74 |
-8.54 |
1 |
2 |
0 |
33 |
261.324 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.90 |
8.2 |
-37.36 |
2 |
2 |
1 |
34 |
262.332 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.44 |
5.68 |
-10.67 |
2 |
3 |
0 |
53 |
277.323 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.44 |
6.14 |
-39.66 |
3 |
3 |
1 |
55 |
278.331 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
C11B1_HUMAN |
P15538
|
Cytochrome P450 11B1, Human |
351 |
0.43 |
Binding ≤ 1μM
|
|
C11B2_HUMAN |
P19099
|
Cytochrome P450 11B2, Human |
110 |
0.46 |
Binding ≤ 1μM
|
|
CP17A_HUMAN |
P05093
|
Cytochrome P450 17A1, Human |
186 |
0.45 |
Binding ≤ 1μM
|
|
CP3A4_HUMAN |
P08684
|
Cytochrome P450 3A4, Human |
896 |
0.40 |
Binding ≤ 1μM
|
|
C11B1_HUMAN |
P15538
|
Cytochrome P450 11B1, Human |
351 |
0.43 |
Binding ≤ 10μM
|
|
C11B2_HUMAN |
P19099
|
Cytochrome P450 11B2, Human |
110 |
0.46 |
Binding ≤ 10μM
|
|
CP17A_HUMAN |
P05093
|
Cytochrome P450 17A1, Human |
186 |
0.45 |
Binding ≤ 10μM
|
|
CP19A_HUMAN |
P11511
|
Cytochrome P450 19A1, Human |
1670 |
0.39 |
Binding ≤ 10μM
|
|
CP3A4_HUMAN |
P08684
|
Cytochrome P450 3A4, Human |
896 |
0.40 |
Binding ≤ 10μM
|
|
CP3A4_HUMAN |
P08684
|
Cytochrome P450 3A4, Human |
896 |
0.40 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.28 |
7.82 |
-9.47 |
1 |
2 |
0 |
33 |
279.314 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.28 |
8.59 |
-49.4 |
0 |
2 |
-1 |
36 |
278.306 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.28 |
8.28 |
-38.99 |
2 |
2 |
1 |
34 |
280.322 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-1-E |
Cytochrome P450 17A1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
343 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.76 |
8.67 |
-8.77 |
1 |
2 |
0 |
33 |
307.368 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.76 |
9.44 |
-48.71 |
0 |
2 |
-1 |
36 |
306.36 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.76 |
9.13 |
-39.07 |
2 |
2 |
1 |
34 |
308.376 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-1-E |
Cytochrome P450 17A1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
343 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.76 |
8.67 |
-8.72 |
1 |
2 |
0 |
33 |
307.368 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.76 |
9.44 |
-48.43 |
0 |
2 |
-1 |
36 |
306.36 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.76 |
9.13 |
-39.72 |
2 |
2 |
1 |
34 |
308.376 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.12 |
11.4 |
-10.97 |
2 |
7 |
0 |
90 |
475.589 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.12 |
11.86 |
-38.75 |
3 |
7 |
1 |
91 |
476.597 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.39 |
10.96 |
-8.97 |
1 |
4 |
0 |
51 |
378.447 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.39 |
11.41 |
-38.5 |
2 |
4 |
1 |
52 |
379.455 |
6 |
↓
|
|
|
|
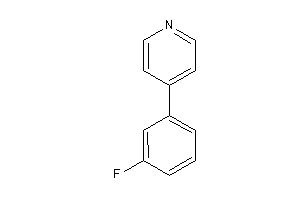
Analogs
-
39320158
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
C11B1-2-E |
Cytochrome P450 11B1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
928 |
0.42 |
Binding ≤ 10μM
|
|
C11B2-1-E |
Cytochrome P450 11B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
515 |
0.44 |
Binding ≤ 10μM
|
|
CP17A-1-E |
Cytochrome P450 17A1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
386 |
0.45 |
Binding ≤ 10μM
|
|
CP3A4-2-E |
Cytochrome P450 3A4 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
668 |
0.43 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.54 |
10.67 |
-7.55 |
0 |
1 |
0 |
13 |
263.315 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.54 |
11.12 |
-36.84 |
1 |
1 |
1 |
14 |
264.323 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
C11B1-2-E |
Cytochrome P450 11B1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4742 |
0.35 |
Binding ≤ 10μM
|
|
C11B2-1-E |
Cytochrome P450 11B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
273 |
0.44 |
Binding ≤ 10μM
|
|
CP17A-1-E |
Cytochrome P450 17A1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
598 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.66 |
10.72 |
-8.54 |
0 |
1 |
0 |
13 |
281.305 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.66 |
11.18 |
-38.86 |
1 |
1 |
1 |
14 |
282.313 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-1-E |
Cytochrome P450 17A1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3340 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.55 |
9.97 |
-9.88 |
0 |
2 |
0 |
22 |
293.341 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
4.55 |
10.43 |
-39.55 |
1 |
2 |
1 |
23 |
294.349 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP17A-1-E |
Cytochrome P450 17A1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1370 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.30 |
10.83 |
-10.19 |
1 |
4 |
0 |
51 |
360.457 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.30 |
11.29 |
-39.04 |
2 |
4 |
1 |
52 |
361.465 |
6 |
↓
|
|