|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.91 |
9.15 |
-10.65 |
3 |
6 |
0 |
95 |
457.622 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
7.23 |
7.51 |
-39.22 |
2 |
6 |
0 |
98 |
456.614 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.48 |
9.23 |
-22.19 |
3 |
6 |
0 |
94 |
458.63 |
3 |
↓
|
|
|
|
Analogs
-
36188745
-
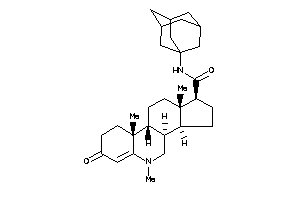
-
36188748
-
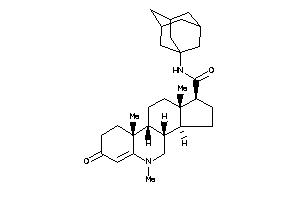
-
60193412
-
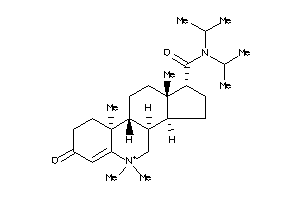
-
60193415
-
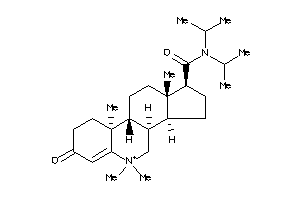
-
60193418
-
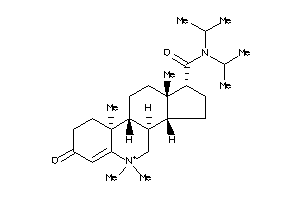
Draw
Identity
99%
90%
80%
70%
Popular Name:
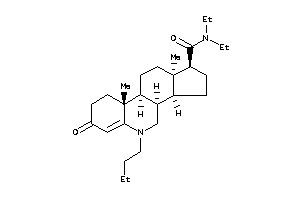
(1S,3aS,3bR,9aR,9bS,11aS)-9a,11a-dimethyl-7-oxo-N-(1,1,3,3-tetramethylbutyl)-1,2,3,3a,3b,4,5,8,9,9b,
(1S,3aS,3bR,9aR,9bS,11aS)-9a,11a…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
3BHS1-2-E |
3-beta-hydroxysteroid Dehydrogenase/delta 5-->4-isomerase Type I (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.31 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
3BHS1_HUMAN |
P14060
|
3-beta-hydroxysteroid Dehydrogenase/delta 5-->4-isomerase Type I, Human |
150 |
0.31 |
Binding ≤ 1μM
|
|
3BHS1_HUMAN |
P14060
|
3-beta-hydroxysteroid Dehydrogenase/delta 5-->4-isomerase Type I, Human |
150 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.56 |
11.51 |
-12.95 |
2 |
4 |
0 |
58 |
428.661 |
4 |
↓
|
|
|
|
Analogs
-
60193476
-
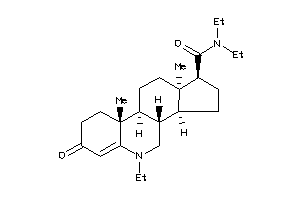
-
60193544
-
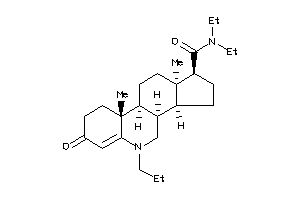
-
60193547
-
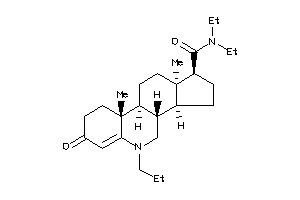
-
60193654
-
-
60193656
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
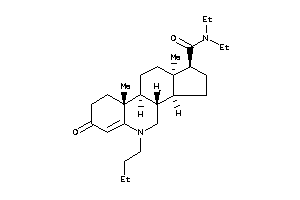
(1S,3aS,3bR,9aR,9bS,11aS)-N-(3,3-dimethylbutyl)-9a,11a-dimethyl-7-oxo-1,2,3,3a,3b,4,5,8,9,9b,10,11-d
(1S,3aS,3bR,9aR,9bS,11aS)-N-(3,3…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
3BHS1-2-E |
3-beta-hydroxysteroid Dehydrogenase/delta 5-->4-isomerase Type I (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8 |
0.39 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
3BHS1_HUMAN |
P14060
|
3-beta-hydroxysteroid Dehydrogenase/delta 5-->4-isomerase Type I, Human |
8.3 |
0.39 |
Binding ≤ 1μM
|
|
3BHS1_HUMAN |
P14060
|
3-beta-hydroxysteroid Dehydrogenase/delta 5-->4-isomerase Type I, Human |
8.3 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.75 |
9.18 |
-14.74 |
2 |
4 |
0 |
58 |
400.607 |
4 |
↓
|
|
|
|
Analogs
-
3916111
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
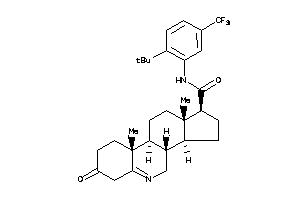
(1S,3aS,3bR,9aR,9bS,11aS)-N-[2-tert-butyl-5-(trifluoromethyl)phenyl]-9a,11a-dimethyl-7-oxo-1,2,3,3a,
(1S,3aS,3bR,9aR,9bS,11aS)-N-[2-t…
Find On:
PubMed —
Wikipedia —
Google
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
3BHS1-2-E |
3-beta-hydroxysteroid Dehydrogenase/delta 5-->4-isomerase Type I (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1600 |
0.22 |
Binding ≤ 10μM
|
|
3BHS2-1-E |
3-beta-hydroxysteroid Dehydrogenase/delta 5-->4-isomerase Type II (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1600 |
0.22 |
Binding ≤ 10μM
|
|
3BHS5-1-E |
3-beta-hydroxysteroid Dehydrogenase Type III (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
140 |
0.26 |
Binding ≤ 10μM
|
|
3BHS7-1-E |
3-beta-hydroxysteroid Dehydrogenase Type VII (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1600 |
0.22 |
Binding ≤ 10μM
|
|
S5A1-1-E |
Steroid 5-alpha-reductase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
S5A2-1-E |
Steroid 5-alpha-reductase 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
3BHS5_RAT |
P27364
|
3-beta-hydroxysteroid Dehydrogenase Type III, Rat |
140 |
0.26 |
Binding ≤ 1μM
|
|
3BHS2_RAT |
P22072
|
3-beta-hydroxysteroid Dehydrogenase/delta 5-->4-isomerase Type II, Rat |
140 |
0.26 |
Binding ≤ 1μM
|
|
S5A1_HUMAN |
P18405
|
Steroid 5-alpha-reductase 1, Human |
8.8 |
0.30 |
Binding ≤ 1μM
|
|
S5A1_RAT |
P24008
|
Steroid 5-alpha-reductase 1, Rat |
0.1 |
0.38 |
Binding ≤ 1μM
|
|
S5A2_RAT |
P31214
|
Steroid 5-alpha-reductase 2, Rat |
0.17 |
0.37 |
Binding ≤ 1μM
|
|
S5A2_HUMAN |
P31213
|
Steroid 5-alpha-reductase 2, Human |
0.1 |
0.38 |
Binding ≤ 1μM
|
|
3BHS5_RAT |
P27364
|
3-beta-hydroxysteroid Dehydrogenase Type III, Rat |
140 |
0.26 |
Binding ≤ 10μM
|
|
3BHS7_HUMAN |
Q9H2F3
|
3-beta-hydroxysteroid Dehydrogenase Type VII, Human |
1600 |
0.22 |
Binding ≤ 10μM
|
|
3BHS1_HUMAN |
P14060
|
3-beta-hydroxysteroid Dehydrogenase/delta 5-->4-isomerase Type I, Human |
1600 |
0.22 |
Binding ≤ 10μM
|
|
3BHS2_HUMAN |
P26439
|
3-beta-hydroxysteroid Dehydrogenase/delta 5-->4-isomerase Type II, Human |
1600 |
0.22 |
Binding ≤ 10μM
|
|
3BHS2_RAT |
P22072
|
3-beta-hydroxysteroid Dehydrogenase/delta 5-->4-isomerase Type II, Rat |
140 |
0.26 |
Binding ≤ 10μM
|
|
S5A1_RAT |
P24008
|
Steroid 5-alpha-reductase 1, Rat |
0.1 |
0.38 |
Binding ≤ 10μM
|
|
S5A1_HUMAN |
P18405
|
Steroid 5-alpha-reductase 1, Human |
8.8 |
0.30 |
Binding ≤ 10μM
|
|
S5A2_RAT |
P31214
|
Steroid 5-alpha-reductase 2, Rat |
0.17 |
0.37 |
Binding ≤ 10μM
|
|
S5A2_HUMAN |
P31213
|
Steroid 5-alpha-reductase 2, Human |
0.1 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.01 |
13.46 |
-17.28 |
2 |
4 |
0 |
58 |
516.648 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.84 |
5.87 |
-43.46 |
0 |
4 |
-1 |
44 |
220.296 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.99 |
4.72 |
-8.39 |
0 |
4 |
0 |
38 |
221.304 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.99 |
5.91 |
-30.71 |
1 |
4 |
1 |
39 |
222.312 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
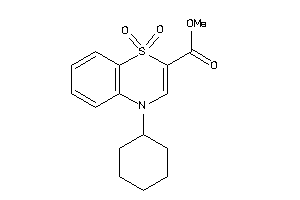
7,8,9,10-tetrahydropyrido[2,1-c][1,4]benzothiazine-6-carbonitrile 5,5-dioxide
7,8,9,10-tetrahydropyrido[2,1-c]…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.21 |
4.68 |
-16.87 |
0 |
4 |
0 |
61 |
260.318 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.64 |
5.33 |
-16.11 |
0 |
4 |
0 |
61 |
274.345 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.64 |
5.34 |
-16.45 |
0 |
4 |
0 |
61 |
274.345 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.35 |
4.74 |
-13.91 |
0 |
4 |
0 |
61 |
278.308 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.35 |
4.73 |
-16.83 |
0 |
4 |
0 |
61 |
278.308 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.87 |
5.18 |
-13.8 |
0 |
4 |
0 |
61 |
294.763 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.87 |
5.18 |
-16.27 |
0 |
4 |
0 |
61 |
294.763 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.00 |
5.28 |
-13.63 |
0 |
4 |
0 |
61 |
339.214 |
0 |
↓
|
|
|
|
Analogs
-
35462512
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.29 |
5.01 |
-16.91 |
0 |
5 |
0 |
64 |
293.344 |
2 |
↓
|
|