|
|
Analogs
-
2017274
-
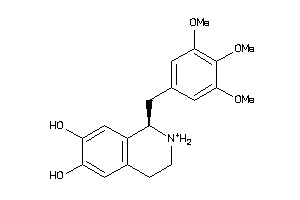
-
33605587
-
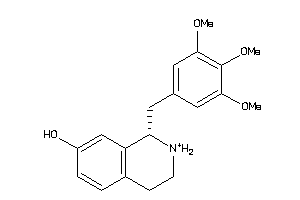
-
33605589
-
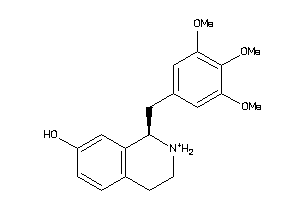
-
38622442
-
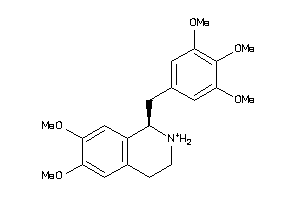
-
38622445
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA1A-3-E |
Alpha-1a Adrenergic Receptor (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.28 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.28 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.28 |
Binding ≤ 10μM
|
|
ADA2A-3-E |
Alpha-2a Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.28 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.28 |
Binding ≤ 10μM
|
|
ADA2C-3-E |
Alpha-2c Adrenergic Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.28 |
Binding ≤ 10μM
|
|
ADRB1-1-E |
Beta-1 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.39 |
Binding ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.39 |
Binding ≤ 10μM
|
|
ADRB3-1-E |
Beta-3 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
100 |
0.39 |
Binding ≤ 10μM
|
|
DRD1-1-E |
Dopamine D1 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5000 |
0.30 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5000 |
0.30 |
Binding ≤ 10μM
|
|
DRD4-2-E |
Dopamine D4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
5000 |
0.30 |
Binding ≤ 10μM
|
|
DRD5-1-E |
Dopamine D5 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.30 |
Binding ≤ 10μM
|
|
ADRB1-1-E |
Beta-1 Adrenergic Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6 |
0.46 |
Functional ≤ 10μM
|
|
ADRB2-1-E |
Beta-2 Adrenergic Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.46 |
Functional ≤ 10μM
|
|
ADRB3-1-E |
Beta-3 Adrenergic Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.49 |
Functional ≤ 10μM
|
|
DRD2-2-E |
Dopamine D2 Receptor (cluster #2 Of 24), Eukaryotic |
Eukaryotes |
5000 |
0.30 |
Binding ≤ 10μM
|
|
Z50587-4-O |
Homo Sapiens (cluster #4 Of 9), Other |
Other |
710 |
0.34 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADRB1_RAT |
P18090
|
Beta-1 Adrenergic Receptor, Rat |
100 |
0.39 |
Binding ≤ 1μM
|
|
ADRB1_HUMAN |
P08588
|
Beta-1 Adrenergic Receptor, Human |
31 |
0.42 |
Binding ≤ 1μM
|
|
ADRB2_CANFA |
P54833
|
Beta-2 Adrenergic Receptor, Canine |
100 |
0.39 |
Binding ≤ 1μM
|
|
ADRB2_HUMAN |
P07550
|
Beta-2 Adrenergic Receptor, Human |
31 |
0.42 |
Binding ≤ 1μM
|
|
ADRB3_RAT |
P26255
|
Beta-3 Adrenergic Receptor, Rat |
100 |
0.39 |
Binding ≤ 1μM
|
|
ADRB3_HUMAN |
P13945
|
Beta-3 Adrenergic Receptor, Human |
31 |
0.42 |
Binding ≤ 1μM
|
|
ADA1A_RAT |
P43140
|
Alpha-1a Adrenergic Receptor, Rat |
10000 |
0.28 |
Binding ≤ 10μM
|
|
ADA1B_RAT |
P15823
|
Alpha-1b Adrenergic Receptor, Rat |
10000 |
0.28 |
Binding ≤ 10μM
|
|
ADA1D_RAT |
P23944
|
Alpha-1d Adrenergic Receptor, Rat |
10000 |
0.28 |
Binding ≤ 10μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
10000 |
0.28 |
Binding ≤ 10μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
10000 |
0.28 |
Binding ≤ 10μM
|
|
ADA2C_RAT |
P22086
|
Alpha-2c Adrenergic Receptor, Rat |
10000 |
0.28 |
Binding ≤ 10μM
|
|
ADRB1_RAT |
P18090
|
Beta-1 Adrenergic Receptor, Rat |
100 |
0.39 |
Binding ≤ 10μM
|
|
ADRB1_HUMAN |
P08588
|
Beta-1 Adrenergic Receptor, Human |
31 |
0.42 |
Binding ≤ 10μM
|
|
ADRB2_HUMAN |
P07550
|
Beta-2 Adrenergic Receptor, Human |
31 |
0.42 |
Binding ≤ 10μM
|
|
ADRB2_CANFA |
P54833
|
Beta-2 Adrenergic Receptor, Canine |
100 |
0.39 |
Binding ≤ 10μM
|
|
ADRB3_HUMAN |
P13945
|
Beta-3 Adrenergic Receptor, Human |
31 |
0.42 |
Binding ≤ 10μM
|
|
ADRB3_RAT |
P26255
|
Beta-3 Adrenergic Receptor, Rat |
100 |
0.39 |
Binding ≤ 10μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
5000 |
0.30 |
Binding ≤ 10μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
5000 |
0.30 |
Binding ≤ 10μM
|
|
DRD3_RAT |
P19020
|
Dopamine D3 Receptor, Rat |
5000 |
0.30 |
Binding ≤ 10μM
|
|
DRD4_RAT |
P30729
|
Dopamine D4 Receptor, Rat |
5000 |
0.30 |
Binding ≤ 10μM
|
|
DRD5_RAT |
P25115
|
Dopamine D5 Receptor, Rat |
5000 |
0.30 |
Binding ≤ 10μM
|
|
ADRB1_HUMAN |
P08588
|
Beta-1 Adrenergic Receptor, Human |
14 |
0.44 |
Functional ≤ 10μM
|
|
ADRB2_HUMAN |
P07550
|
Beta-2 Adrenergic Receptor, Human |
3.5 |
0.47 |
Functional ≤ 10μM
|
|
ADRB3_HUMAN |
P13945
|
Beta-3 Adrenergic Receptor, Human |
1.7 |
0.49 |
Functional ≤ 10μM
|
|
Z50587 |
Z50587
|
Homo Sapiens |
550 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
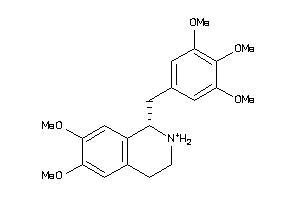
2.12 |
-2.75 |
-54.23 |
4 |
6 |
1 |
84 |
346.403 |
5 |
↓
|
|
|
|
|
|
|
Analogs
-
14262043
-
-
14262045
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SOAT1-1-E |
Acyl Coenzyme A:cholesterol Acyltransferase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2700 |
0.18 |
Binding ≤ 10μM |
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SOAT1_RAT |
O70536
|
Acyl Coenzyme A:cholesterol Acyltransferase 1, Rat |
2700 |
0.18 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
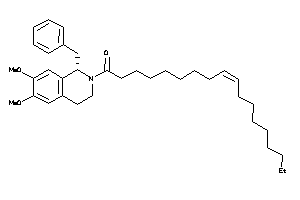
9.21 |
21.04 |
-18.37 |
0 |
6 |
0 |
57 |
607.876 |
21 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SOAT1-1-E |
Acyl Coenzyme A:cholesterol Acyltransferase 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2700 |
0.18 |
Binding ≤ 10μM |
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SOAT1_RAT |
O70536
|
Acyl Coenzyme A:cholesterol Acyltransferase 1, Rat |
2700 |
0.18 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
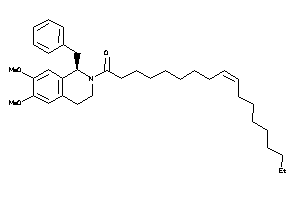
9.21 |
19.45 |
-14.39 |
0 |
6 |
0 |
57 |
607.876 |
21 |
↓
|
|
|
|
Analogs
-
15263003
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
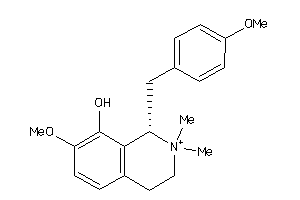
2.60 |
5.51 |
-43.7 |
3 |
5 |
1 |
63 |
330.404 |
4 |
↓
|
|
|
|
Analogs
-
15263003
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.37 |
6.4 |
-36.05 |
2 |
5 |
1 |
59 |
344.431 |
4 |
↓
|
|
|
|
Analogs
-
15263003
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.37 |
6.4 |
-36.22 |
2 |
5 |
1 |
59 |
344.431 |
4 |
↓
|
|
|
|
|