|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.02 |
6.75 |
-101.75 |
5 |
7 |
2 |
88 |
526.505 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.02 |
3.1 |
-12.16 |
3 |
7 |
0 |
85 |
524.489 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.02 |
4.69 |
-53.1 |
4 |
7 |
1 |
86 |
525.497 |
9 |
↓
|
|
|
|
Analogs
-
36215760
-
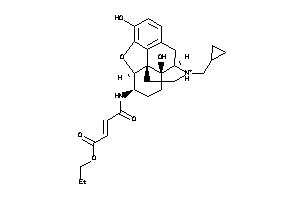
-
2539826
-
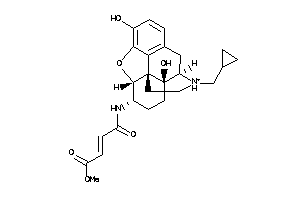
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD_RAT |
P33533
|
Delta Opioid Receptor, Rat |
1.03 |
0.38 |
Binding ≤ 1μM
|
|
OPRK_RAT |
P34975
|
Kappa Opioid Receptor, Rat |
1.03 |
0.38 |
Binding ≤ 1μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
1.03 |
0.38 |
Binding ≤ 1μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
1.03 |
0.38 |
Binding ≤ 1μM
|
|
OPRD_RAT |
P33533
|
Delta Opioid Receptor, Rat |
1.03 |
0.38 |
Binding ≤ 10μM
|
|
OPRK_RAT |
P34975
|
Kappa Opioid Receptor, Rat |
1.03 |
0.38 |
Binding ≤ 10μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
1.03 |
0.38 |
Binding ≤ 10μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
1.03 |
0.38 |
Binding ≤ 10μM
|
|
OPRD_MOUSE |
P32300
|
Delta Opioid Receptor, Mouse |
82 |
0.30 |
Functional ≤ 10μM
|
|
OPRK_MOUSE |
P33534
|
Kappa Opioid Receptor, Mouse |
82 |
0.30 |
Functional ≤ 10μM
|
|
OPRM_MOUSE |
P42866
|
Mu Opioid Receptor, Mouse |
82 |
0.30 |
Functional ≤ 10μM
|
|
SGMR1_MOUSE |
O55242
|
Sigma Opioid Receptor, Mouse |
82 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.65 |
5.71 |
-62.47 |
4 |
8 |
1 |
110 |
455.531 |
6 |
↓
|
|
|
|
Analogs
-
36215763
-
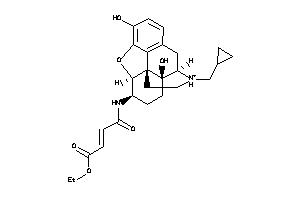
-
36215765
-
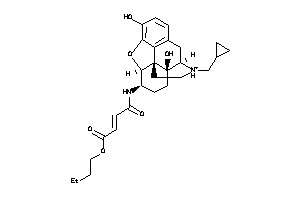
-
36215776
-
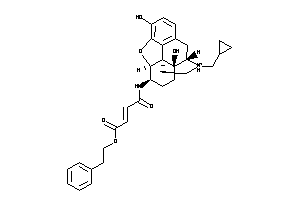
-
36215780
-
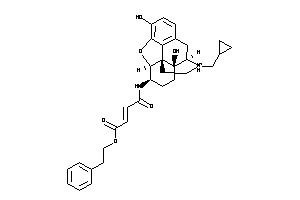
-
29544392
-
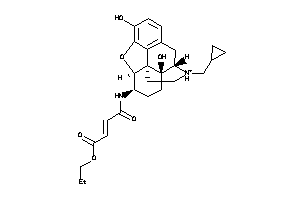
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD_RAT |
P33533
|
Delta Opioid Receptor, Rat |
1.56 |
0.35 |
Binding ≤ 1μM
|
|
OPRK_RAT |
P34975
|
Kappa Opioid Receptor, Rat |
1.56 |
0.35 |
Binding ≤ 1μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
1.56 |
0.35 |
Binding ≤ 1μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
1.56 |
0.35 |
Binding ≤ 1μM
|
|
OPRD_RAT |
P33533
|
Delta Opioid Receptor, Rat |
1.56 |
0.35 |
Binding ≤ 10μM
|
|
OPRK_RAT |
P34975
|
Kappa Opioid Receptor, Rat |
1.56 |
0.35 |
Binding ≤ 10μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
1.56 |
0.35 |
Binding ≤ 10μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
1.56 |
0.35 |
Binding ≤ 10μM
|
|
OPRD_MOUSE |
P32300
|
Delta Opioid Receptor, Mouse |
21 |
0.31 |
Functional ≤ 10μM
|
|
OPRK_MOUSE |
P33534
|
Kappa Opioid Receptor, Mouse |
21 |
0.31 |
Functional ≤ 10μM
|
|
OPRM_MOUSE |
P42866
|
Mu Opioid Receptor, Mouse |
21 |
0.31 |
Functional ≤ 10μM
|
|
SGMR1_MOUSE |
O55242
|
Sigma Opioid Receptor, Mouse |
21 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.53 |
7.44 |
-61.94 |
4 |
8 |
1 |
110 |
483.585 |
8 |
↓
|
|
|
|
Analogs
-
36215765
-

-
36215776
-

-
36215780
-

-
36215760
-

Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD_RAT |
P33533
|
Delta Opioid Receptor, Rat |
0.64 |
0.38 |
Binding ≤ 1μM
|
|
OPRK_RAT |
P34975
|
Kappa Opioid Receptor, Rat |
0.64 |
0.38 |
Binding ≤ 1μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
0.64 |
0.38 |
Binding ≤ 1μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
0.64 |
0.38 |
Binding ≤ 1μM
|
|
OPRD_RAT |
P33533
|
Delta Opioid Receptor, Rat |
0.64 |
0.38 |
Binding ≤ 10μM
|
|
OPRK_RAT |
P34975
|
Kappa Opioid Receptor, Rat |
0.64 |
0.38 |
Binding ≤ 10μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
0.64 |
0.38 |
Binding ≤ 10μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
0.64 |
0.38 |
Binding ≤ 10μM
|
|
OPRD_MOUSE |
P32300
|
Delta Opioid Receptor, Mouse |
13 |
0.32 |
Functional ≤ 10μM
|
|
OPRK_MOUSE |
P33534
|
Kappa Opioid Receptor, Mouse |
13 |
0.32 |
Functional ≤ 10μM
|
|
OPRM_MOUSE |
P42866
|
Mu Opioid Receptor, Mouse |
13 |
0.32 |
Functional ≤ 10μM
|
|
SGMR1_MOUSE |
O55242
|
Sigma Opioid Receptor, Mouse |
13 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.03 |
6.64 |
-62.16 |
4 |
8 |
1 |
110 |
469.558 |
7 |
↓
|
|
|
|
Analogs
-
36215776
-

-
36215780
-

-
36215760
-

Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD_RAT |
P33533
|
Delta Opioid Receptor, Rat |
1.65 |
0.34 |
Binding ≤ 1μM
|
|
OPRK_RAT |
P34975
|
Kappa Opioid Receptor, Rat |
1.65 |
0.34 |
Binding ≤ 1μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
1.65 |
0.34 |
Binding ≤ 1μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
1.65 |
0.34 |
Binding ≤ 1μM
|
|
OPRD_RAT |
P33533
|
Delta Opioid Receptor, Rat |
1.65 |
0.34 |
Binding ≤ 10μM
|
|
OPRK_RAT |
P34975
|
Kappa Opioid Receptor, Rat |
1.65 |
0.34 |
Binding ≤ 10μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
1.65 |
0.34 |
Binding ≤ 10μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
1.65 |
0.34 |
Binding ≤ 10μM
|
|
OPRD_MOUSE |
P32300
|
Delta Opioid Receptor, Mouse |
7 |
0.32 |
Functional ≤ 10μM
|
|
OPRK_MOUSE |
P33534
|
Kappa Opioid Receptor, Mouse |
7 |
0.32 |
Functional ≤ 10μM
|
|
OPRM_MOUSE |
P42866
|
Mu Opioid Receptor, Mouse |
7 |
0.32 |
Functional ≤ 10μM
|
|
SGMR1_MOUSE |
O55242
|
Sigma Opioid Receptor, Mouse |
7 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.09 |
8.22 |
-61.9 |
4 |
8 |
1 |
110 |
497.612 |
9 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.05 |
6.45 |
-58.51 |
3 |
7 |
1 |
84 |
415.51 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.05 |
4.72 |
-12.63 |
2 |
7 |
0 |
82 |
414.502 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.52 |
3.9 |
-58.09 |
3 |
7 |
1 |
98 |
422.523 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.52 |
2.18 |
-14.05 |
2 |
7 |
0 |
96 |
421.515 |
4 |
↓
|
|