|
|
Analogs
-
3950170
-
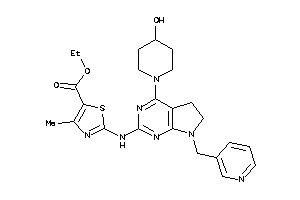
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE7A-1-E |
Phosphodiesterase 7A (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.18 |
Binding ≤ 10μM
|
|
PDE7B-1-E |
Phosphodiesterase 7B (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.18 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.01 |
13.99 |
-114.38 |
4 |
12 |
2 |
129 |
571.704 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.01 |
12.69 |
-40.53 |
3 |
12 |
1 |
124 |
570.696 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.88 |
6.21 |
-15.01 |
0 |
5 |
0 |
63 |
327.768 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PK3CA_HUMAN |
P42336
|
PI3-kinase P110-alpha Subunit, Human |
14 |
0.42 |
Binding ≤ 1μM
|
|
PK3CB_HUMAN |
P42338
|
PI3-kinase P110-beta Subunit, Human |
120 |
0.37 |
Binding ≤ 1μM
|
|
PK3CD_HUMAN |
O00329
|
PI3-kinase P110-delta Subunit, Human |
500 |
0.34 |
Binding ≤ 1μM
|
|
PK3CG_HUMAN |
P48736
|
PI3-kinase P110-gamma Subunit, Human |
36 |
0.40 |
Binding ≤ 1μM
|
|
MTOR_HUMAN |
P42345
|
FK506 Binding Protein 12, Human |
1600 |
0.31 |
Binding ≤ 10μM
|
|
P3C2B_HUMAN |
O00750
|
Phosphatidylinositol-4-phosphate 3-kinase C2 Domain-containing Beta Polypeptide, Human |
5300 |
0.28 |
Binding ≤ 10μM
|
|
PK3CA_HUMAN |
P42336
|
PI3-kinase P110-alpha Subunit, Human |
14 |
0.42 |
Binding ≤ 10μM
|
|
PK3CB_HUMAN |
P42338
|
PI3-kinase P110-beta Subunit, Human |
120 |
0.37 |
Binding ≤ 10μM
|
|
PK3CD_HUMAN |
O00329
|
PI3-kinase P110-delta Subunit, Human |
500 |
0.34 |
Binding ≤ 10μM
|
|
PK3CG_HUMAN |
P48736
|
PI3-kinase P110-gamma Subunit, Human |
36 |
0.40 |
Binding ≤ 10μM
|
|
Z80928 |
Z80928
|
HCT-116 (Colon Carcinoma Cells) |
200 |
0.36 |
Functional ≤ 10μM
|
|
Z80493 |
Z80493
|
SK-OV-3 (Ovarian Carcinoma Cells) |
120 |
0.37 |
Functional ≤ 10μM
|
|
Z80712 |
Z80712
|
T47D (Breast Carcinoma Cells) |
56 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.30 |
5.16 |
-16.72 |
2 |
10 |
0 |
127 |
377.43 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PK3CA-2-E |
PI3-kinase P110-alpha Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
77 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.57 |
10.91 |
-35.39 |
3 |
8 |
1 |
92 |
415.477 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.57 |
10.62 |
-17.21 |
2 |
8 |
0 |
90 |
414.469 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.57 |
11.39 |
-79.88 |
4 |
8 |
2 |
93 |
416.485 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.86 |
7.95 |
-126.76 |
7 |
12 |
-1 |
204 |
456.483 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.86 |
5.98 |
-78.75 |
8 |
12 |
0 |
201 |
457.491 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.86 |
7.99 |
-128.11 |
7 |
12 |
-1 |
204 |
456.483 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.86 |
6.02 |
-79.46 |
8 |
12 |
0 |
201 |
457.491 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.11 |
6.2 |
-128.17 |
8 |
12 |
-1 |
212 |
442.456 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.11 |
4.22 |
-81.62 |
9 |
12 |
0 |
210 |
443.464 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.11 |
6.2 |
-128.12 |
8 |
12 |
-1 |
212 |
442.456 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.11 |
4.22 |
-82.07 |
9 |
12 |
0 |
210 |
443.464 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.72 |
9.17 |
-125.05 |
7 |
11 |
-1 |
200 |
455.495 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.72 |
8.71 |
-118.9 |
6 |
11 |
-2 |
199 |
454.487 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.72 |
7.19 |
-76.71 |
8 |
11 |
0 |
198 |
456.503 |
10 |
↓
|
|