|
|
Analogs
-
34849740
-
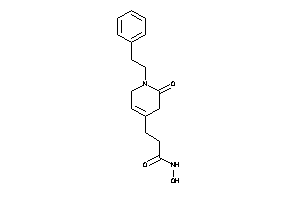
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDA10-2-E |
Histone Deacetylase 10 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
350 |
0.43 |
Binding ≤ 10μM
|
|
HDA11-3-E |
Histone Deacetylase 11 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
350 |
0.43 |
Binding ≤ 10μM
|
|
HDAC1-1-E |
Histone Deacetylase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
350 |
0.43 |
Binding ≤ 10μM
|
|
HDAC2-1-E |
Histone Deacetylase 2 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
350 |
0.43 |
Binding ≤ 10μM
|
|
HDAC3-2-E |
Histone Deacetylase 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
350 |
0.43 |
Binding ≤ 10μM
|
|
HDAC4-2-E |
Histone Deacetylase 4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
350 |
0.43 |
Binding ≤ 10μM
|
|
HDAC5-2-E |
Histone Deacetylase 5 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
350 |
0.43 |
Binding ≤ 10μM
|
|
HDAC6-2-E |
Histone Deacetylase 6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
350 |
0.43 |
Binding ≤ 10μM
|
|
HDAC7-3-E |
Histone Deacetylase 7 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
350 |
0.43 |
Binding ≤ 10μM
|
|
HDAC8-1-E |
Histone Deacetylase 8 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
350 |
0.43 |
Binding ≤ 10μM
|
|
HDAC9-3-E |
Histone Deacetylase 9 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
350 |
0.43 |
Binding ≤ 10μM
|
|
Z80418-9-O |
RAW264.7 (Monocytic-macrophage Leukemia Cells) (cluster #9 Of 9), Other |
Other |
4270 |
0.36 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HDAC1_HUMAN |
Q13547
|
Histone Deacetylase 1, Human |
350 |
0.43 |
Binding ≤ 1μM
|
|
HDA10_HUMAN |
Q969S8
|
Histone Deacetylase 10, Human |
350 |
0.43 |
Binding ≤ 1μM
|
|
HDA11_HUMAN |
Q96DB2
|
Histone Deacetylase 11, Human |
350 |
0.43 |
Binding ≤ 1μM
|
|
HDAC2_HUMAN |
Q92769
|
Histone Deacetylase 2, Human |
350 |
0.43 |
Binding ≤ 1μM
|
|
HDAC3_HUMAN |
O15379
|
Histone Deacetylase 3, Human |
350 |
0.43 |
Binding ≤ 1μM
|
|
HDAC4_HUMAN |
P56524
|
Histone Deacetylase 4, Human |
350 |
0.43 |
Binding ≤ 1μM
|
|
HDAC5_HUMAN |
Q9UQL6
|
Histone Deacetylase 5, Human |
350 |
0.43 |
Binding ≤ 1μM
|
|
HDAC6_HUMAN |
Q9UBN7
|
Histone Deacetylase 6, Human |
350 |
0.43 |
Binding ≤ 1μM
|
|
HDAC7_HUMAN |
Q8WUI4
|
Histone Deacetylase 7, Human |
350 |
0.43 |
Binding ≤ 1μM
|
|
HDAC8_HUMAN |
Q9BY41
|
Histone Deacetylase 8, Human |
350 |
0.43 |
Binding ≤ 1μM
|
|
HDAC9_HUMAN |
Q9UKV0
|
Histone Deacetylase 9, Human |
350 |
0.43 |
Binding ≤ 1μM
|
|
HDAC1_HUMAN |
Q13547
|
Histone Deacetylase 1, Human |
350 |
0.43 |
Binding ≤ 10μM
|
|
HDA10_HUMAN |
Q969S8
|
Histone Deacetylase 10, Human |
350 |
0.43 |
Binding ≤ 10μM
|
|
HDA11_HUMAN |
Q96DB2
|
Histone Deacetylase 11, Human |
350 |
0.43 |
Binding ≤ 10μM
|
|
HDAC2_HUMAN |
Q92769
|
Histone Deacetylase 2, Human |
350 |
0.43 |
Binding ≤ 10μM
|
|
HDAC3_HUMAN |
O15379
|
Histone Deacetylase 3, Human |
350 |
0.43 |
Binding ≤ 10μM
|
|
HDAC4_HUMAN |
P56524
|
Histone Deacetylase 4, Human |
350 |
0.43 |
Binding ≤ 10μM
|
|
HDAC5_HUMAN |
Q9UQL6
|
Histone Deacetylase 5, Human |
350 |
0.43 |
Binding ≤ 10μM
|
|
HDAC6_HUMAN |
Q9UBN7
|
Histone Deacetylase 6, Human |
350 |
0.43 |
Binding ≤ 10μM
|
|
HDAC7_HUMAN |
Q8WUI4
|
Histone Deacetylase 7, Human |
350 |
0.43 |
Binding ≤ 10μM
|
|
HDAC8_HUMAN |
Q9BY41
|
Histone Deacetylase 8, Human |
350 |
0.43 |
Binding ≤ 10μM
|
|
HDAC9_HUMAN |
Q9UKV0
|
Histone Deacetylase 9, Human |
350 |
0.43 |
Binding ≤ 10μM
|
|
Z80418 |
Z80418
|
RAW264.7 (Monocytic-macrophage Leukemia Cells) |
1900 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.39 |
6.06 |
-20.51 |
2 |
5 |
0 |
70 |
288.347 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.39 |
7.27 |
-65.89 |
1 |
5 |
-1 |
72 |
287.339 |
6 |
↓
|
|