|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.18 |
4.01 |
-48.87 |
0 |
4 |
-1 |
58 |
139.134 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.18 |
4.53 |
-50.01 |
1 |
4 |
0 |
59 |
140.142 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
-
3650249
-
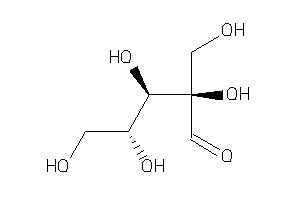
-
8552145
-
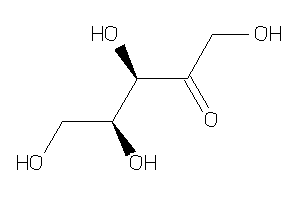
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.22 |
-7.48 |
-11.99 |
4 |
5 |
0 |
98 |
150.13 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
-2.22 |
-7.36 |
-11.73 |
4 |
5 |
0 |
98 |
150.13 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
-2.22 |
-5.76 |
-11.37 |
4 |
5 |
0 |
98 |
150.13 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.42 |
4.8 |
-137.63 |
6 |
13 |
-2 |
208 |
457.447 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.42 |
4.84 |
-142.05 |
7 |
13 |
-1 |
210 |
458.455 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.42 |
2.92 |
-94.98 |
8 |
13 |
0 |
207 |
459.463 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5NTD-1-E |
5'-nucleotidase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1 |
0.47 |
Binding ≤ 10μM
|
|
GRP78-1-E |
78 KDa Glucose-regulated Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3830 |
0.28 |
Binding ≤ 10μM
|
|
HSP7C-1-E |
Heat Shock Cognate 71 KDa Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
110 |
0.36 |
Binding ≤ 10μM
|
|
KAP2-1-E |
CAMP-dependent Protein Kinase Type II-alpha Regulatory Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.26 |
Binding ≤ 10μM
|
|
KAPCA-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.26 |
Binding ≤ 10μM
|
|
KAPCB-1-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.26 |
Binding ≤ 10μM
|
|
P2RX1-1-E |
P2X Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.26 |
Functional ≤ 10μM
|
|
P2RY1-1-E |
P2Y Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
100 |
0.36 |
Functional ≤ 10μM
|
|
P2Y12-1-E |
Purinergic Receptor P2Y12 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
69 |
0.37 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5NTD_RAT |
P21588
|
5'-nucleotidase, Rat |
0.91 |
0.47 |
Binding ≤ 1μM
|
|
HSP7C_BOVIN |
P19120
|
Heat Shock Cognate 71 KDa Protein, Bovin |
110 |
0.36 |
Binding ≤ 1μM
|
|
HSP7C_HUMAN |
P11142
|
Heat Shock Cognate 71 KDa Protein, Human |
260 |
0.34 |
Binding ≤ 1μM
|
|
5NTD_RAT |
P21588
|
5'-nucleotidase, Rat |
0.91 |
0.47 |
Binding ≤ 10μM
|
|
GRP78_HUMAN |
P11021
|
78 KDa Glucose-regulated Protein, Human |
2170 |
0.29 |
Binding ≤ 10μM
|
|
KAPCA_MOUSE |
P05132
|
CAMP-dependent Protein Kinase Alpha-catalytic Subunit, Mouse |
10000 |
0.26 |
Binding ≤ 10μM
|
|
KAPCB_MOUSE |
P68181
|
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit, Mouse |
10000 |
0.26 |
Binding ≤ 10μM
|
|
KAP2_BOVIN |
P00515
|
CAMP-dependent Protein Kinase Type II-alpha Regulatory Subunit, Bovin |
10000 |
0.26 |
Binding ≤ 10μM
|
|
HSP7C_BOVIN |
P19120
|
Heat Shock Cognate 71 KDa Protein, Bovin |
110 |
0.36 |
Binding ≤ 10μM
|
|
HSP7C_HUMAN |
P11142
|
Heat Shock Cognate 71 KDa Protein, Human |
260 |
0.34 |
Binding ≤ 10μM
|
|
P2RX1_HUMAN |
P51575
|
P2X Purinoceptor 1, Human |
10000 |
0.26 |
Functional ≤ 10μM
|
|
P2RY1_MELGA |
P49652
|
P2Y Purinoceptor 1, Melga |
100 |
0.36 |
Functional ≤ 10μM
|
|
P2RY1_HUMAN |
P47900
|
Purinergic Receptor P2Y1, Human |
14 |
0.41 |
Functional ≤ 10μM
|
|
P2Y12_RAT |
Q9EPX4
|
Purinergic Receptor P2Y12, Rat |
69 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Hi
High (pH 8-9.5)
|
3.79 |
9.28 |
-36.43 |
1 |
2 |
1 |
8 |
233.379 |
3 |
↓
|
|
|
|
Analogs
-
12402869
-
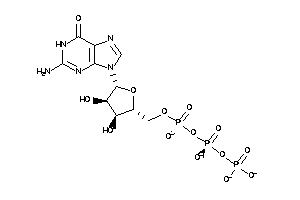
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.22 |
-7.52 |
-180.6 |
6 |
19 |
-3 |
308 |
520.157 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.53 |
-4.25 |
-237.08 |
6 |
19 |
-3 |
308 |
520.157 |
8 |
↓
|
|
|
|
|
|
|
Analogs
-
4228257
-
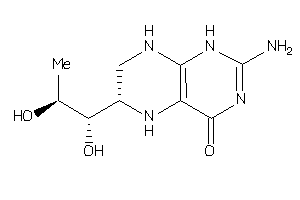
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1-1-E |
Nitric-oxide Synthase, Brain (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
1120 |
0.49 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS1_HUMAN |
P29475
|
Nitric-oxide Synthase, Brain, Human |
1120 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.15 |
-4.44 |
-11.69 |
7 |
8 |
0 |
136 |
241.251 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
-1.15 |
-5.1 |
-20.07 |
7 |
8 |
0 |
136 |
241.251 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KAP2-1-E |
CAMP-dependent Protein Kinase Type II-alpha Regulatory Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.23 |
Binding ≤ 10μM
|
|
KAPCA-1-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.23 |
Binding ≤ 10μM
|
|
KAPCB-1-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10000 |
0.23 |
Binding ≤ 10μM
|
|
MAPK2-1-E |
MAP Kinase-activated Protein Kinase 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6300 |
0.23 |
Binding ≤ 10μM
|
|
P2RY1-1-E |
Purinergic Receptor P2Y1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2800 |
0.25 |
Binding ≤ 10μM
|
|
P2RX1-1-E |
P2X Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
56 |
0.33 |
Functional ≤ 10μM
|
|
P2RX2-1-E |
P2X Purinoceptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.26 |
Functional ≤ 10μM
|
|
P2RX3-1-E |
P2X Purinoceptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
340 |
0.29 |
Functional ≤ 10μM
|
|
P2RX4-1-E |
P2X Purinoceptor 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.28 |
Functional ≤ 10μM
|
|
P2RX5-1-E |
P2X Purinoceptor 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.27 |
Functional ≤ 10μM
|
|
P2RX6-1-E |
P2X Purinoceptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.27 |
Functional ≤ 10μM
|
|
P2RY1-1-E |
P2Y Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7200 |
0.23 |
Functional ≤ 10μM
|
|
P2RY2-1-E |
Purinergic Receptor P2Y2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
96 |
0.32 |
Functional ≤ 10μM
|
|
P2RY4-1-E |
Pyrimidinergic Receptor P2Y4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1800 |
0.26 |
Functional ≤ 10μM
|
|
P2Y11-1-E |
Purinergic Receptor P2Y11 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3300 |
0.25 |
Functional ≤ 10μM |
|
P2Y12-1-E |
Purinergic Receptor P2Y12 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3190 |
0.25 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KAPCA_MOUSE |
P05132
|
CAMP-dependent Protein Kinase Alpha-catalytic Subunit, Mouse |
10000 |
0.23 |
Binding ≤ 10μM
|
|
KAPCB_MOUSE |
P68181
|
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit, Mouse |
10000 |
0.23 |
Binding ≤ 10μM
|
|
KAP2_BOVIN |
P00515
|
CAMP-dependent Protein Kinase Type II-alpha Regulatory Subunit, Bovin |
10000 |
0.23 |
Binding ≤ 10μM
|
|
MAPK2_HUMAN |
P49137
|
MAP Kinase-activated Protein Kinase 2, Human |
2800 |
0.25 |
Binding ≤ 10μM
|
|
P2RY1_HUMAN |
P47900
|
Purinergic Receptor P2Y1, Human |
1500 |
0.26 |
Binding ≤ 10μM
|
|
P2RX1_HUMAN |
P51575
|
P2X Purinoceptor 1, Human |
56 |
0.33 |
Functional ≤ 10μM
|
|
P2RX2_RAT |
P49653
|
P2X Purinoceptor 2, Rat |
1400 |
0.26 |
Functional ≤ 10μM
|
|
P2RX3_HUMAN |
P56373
|
P2X Purinoceptor 3, Human |
340 |
0.29 |
Functional ≤ 10μM
|
|
P2RX4_RAT |
P51577
|
P2X Purinoceptor 4, Rat |
1000 |
0.27 |
Functional ≤ 10μM
|
|
P2RX4_HUMAN |
Q99571
|
P2X Purinoceptor 4, Human |
500 |
0.28 |
Functional ≤ 10μM
|
|
P2RX5_RAT |
P51578
|
P2X Purinoceptor 5, Rat |
1000 |
0.27 |
Functional ≤ 10μM
|
|
P2RX6_RAT |
P51579
|
P2X Purinoceptor 6, Rat |
1000 |
0.27 |
Functional ≤ 10μM
|
|
P2RY1_HUMAN |
P47900
|
Purinergic Receptor P2Y1, Human |
14 |
0.35 |
Functional ≤ 10μM
|
|
P2Y11_HUMAN |
Q96G91
|
Purinergic Receptor P2Y11, Human |
3300 |
0.25 |
Functional ≤ 10μM |
|
P2Y12_RAT |
Q9EPX4
|
Purinergic Receptor P2Y12, Rat |
3190 |
0.25 |
Functional ≤ 10μM
|
|
P2RY2_HUMAN |
P41231
|
Purinergic Receptor P2Y2, Human |
230 |
0.30 |
Functional ≤ 10μM
|
|
P2RY4_RAT |
O35811
|
Pyrimidinergic Receptor P2Y4, Rat |
1800 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RY1-1-E |
P2Y Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.39 |
Functional ≤ 10μM
|
|
P2RY1-1-E |
P2Y Purinoceptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
590 |
0.28 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.93 |
-9.04 |
-49.11 |
4 |
6 |
-1 |
117 |
175.116 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.12 |
-4.63 |
-15.58 |
2 |
6 |
0 |
107 |
175.116 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.12 |
-4.85 |
-15.15 |
2 |
6 |
0 |
107 |
175.116 |
2 |
↓
|
|
|
|
|
|
|
Analogs
-
4228257
-

-
4228258
-
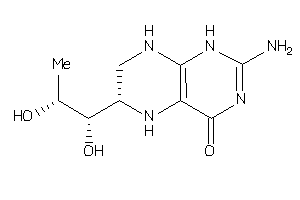
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.15 |
-5.25 |
-21.45 |
7 |
8 |
0 |
136 |
241.251 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.63 |
9.32 |
-60.23 |
0 |
5 |
0 |
66 |
245.319 |
9 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.71 |
11.19 |
-45.24 |
1 |
3 |
-1 |
60 |
295.443 |
14 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.71 |
9.21 |
-6.87 |
2 |
3 |
0 |
58 |
296.451 |
14 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.08 |
14.2 |
-48.93 |
0 |
3 |
-1 |
57 |
317.449 |
14 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.08 |
12.23 |
-10.83 |
1 |
3 |
0 |
54 |
318.457 |
14 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.43 |
24.67 |
-56.34 |
2 |
8 |
0 |
108 |
703.043 |
36 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
5851414
-
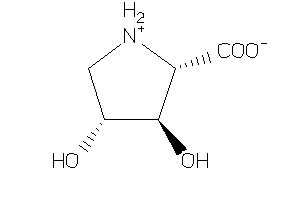
-
36466335
-
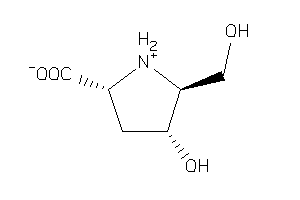
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.64 |
-4.33 |
-44.14 |
3 |
4 |
0 |
76 |
131.131 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.50 |
7.8 |
-23.77 |
2 |
9 |
0 |
104 |
505.497 |
7 |
↓
|
|
|
|
Analogs
-
5851414
-

-
36466335
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.64 |
-4.23 |
-42.31 |
3 |
4 |
0 |
76 |
131.131 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.89 |
10.67 |
-48.01 |
1 |
3 |
-1 |
60 |
319.465 |
15 |
↓
|
|
|
|
Analogs
-
2516113
-
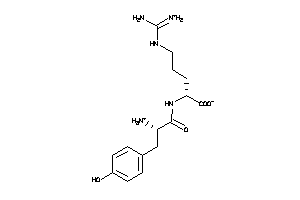
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.21 |
0.73 |
-96.18 |
10 |
9 |
1 |
181 |
338.388 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.21 |
0.46 |
-67.06 |
9 |
9 |
0 |
179 |
337.38 |
10 |
↓
|
|
|
|
Analogs
-
4262577
-
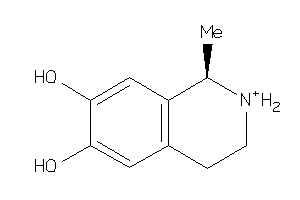
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD4-2-E |
Dopamine D4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6400 |
0.56 |
Binding ≤ 10μM |
|
DRD2-2-E |
Dopamine D2 Receptor (cluster #2 Of 24), Eukaryotic |
Eukaryotes |
5400 |
0.57 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.06 |
0.19 |
-45.11 |
4 |
3 |
1 |
57 |
180.227 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.06 |
-1.12 |
-6.08 |
3 |
3 |
0 |
52 |
179.219 |
0 |
↓
|
|
|
|
Analogs
-
4027288
-
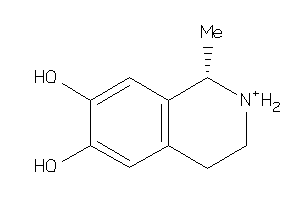
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRD4-2-E |
Dopamine D4 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
6400 |
0.56 |
Binding ≤ 10μM |
|
DRD2-2-E |
Dopamine D2 Receptor (cluster #2 Of 24), Eukaryotic |
Eukaryotes |
5400 |
0.57 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.06 |
0.2 |
-45.49 |
4 |
3 |
1 |
57 |
180.227 |
0 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|