|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.11 |
4.6 |
-6.99 |
0 |
5 |
0 |
70 |
253.048 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.11 |
4.72 |
-7 |
0 |
5 |
0 |
70 |
253.048 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.54 |
5.65 |
-38.45 |
1 |
5 |
0 |
79 |
269.054 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.84 |
5.23 |
-38.14 |
0 |
5 |
-1 |
82 |
268.046 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACH10-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
224 |
0.85 |
Binding ≤ 10μM |
|
ACHA3-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
224 |
0.85 |
Binding ≤ 10μM |
|
ACHA4-5-E |
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
224 |
0.85 |
Binding ≤ 10μM |
|
ACM1-4-E |
Muscarinic Acetylcholine Receptor M1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
5 |
1.06 |
Binding ≤ 10μM
|
|
ACM2-6-E |
Muscarinic Acetylcholine Receptor M2 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
9150 |
0.64 |
Binding ≤ 10μM |
|
ACM3-4-E |
Muscarinic Acetylcholine Receptor M3 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
5700 |
0.67 |
Binding ≤ 10μM
|
|
ACM4-6-E |
Muscarinic Acetylcholine Receptor M4 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
7 |
1.04 |
Binding ≤ 10μM
|
|
ACM5-3-E |
Muscarinic Acetylcholine Receptor M5 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
600 |
0.79 |
Binding ≤ 10μM
|
|
ACM1-1-E |
Muscarinic Acetylcholine Receptor M1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
73 |
0.91 |
Functional ≤ 10μM
|
|
ACM2-3-E |
Muscarinic Acetylcholine Receptor M2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
73 |
0.91 |
Functional ≤ 10μM
|
|
ACM3-3-E |
Muscarinic Acetylcholine Receptor M3 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
73 |
0.91 |
Functional ≤ 10μM
|
|
ACM4-3-E |
Muscarinic Acetylcholine Receptor M4 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
73 |
0.91 |
Functional ≤ 10μM
|
|
ACM5-3-E |
Muscarinic Acetylcholine Receptor M5 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
73 |
0.91 |
Functional ≤ 10μM
|
|
Z104290-3-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #3 Of 4), Other |
Other |
270 |
0.84 |
Binding ≤ 10μM
|
|
Z104303-5-O |
Muscarinic Acetylcholine Receptor (cluster #5 Of 7), Other |
Other |
77 |
0.91 |
Binding ≤ 10μM |
|
Z50512-4-O |
Cavia Porcellus (cluster #4 Of 7), Other |
Other |
600 |
0.79 |
Functional ≤ 10μM
|
|
Z50592-8-O |
Oryctolagus Cuniculus (cluster #8 Of 8), Other |
Other |
545 |
0.80 |
Functional ≤ 10μM |
|
Z80024-2-O |
A9 (Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
1200 |
0.75 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104303 |
Z104303
|
Muscarinic Acetylcholine Receptor |
1.9 |
1.11 |
Binding ≤ 1μM
|
|
ACM1_MOUSE |
P12657
|
Muscarinic Acetylcholine Receptor M1, Mouse |
3.33 |
1.08 |
Binding ≤ 1μM
|
|
ACM1_HUMAN |
P11229
|
Muscarinic Acetylcholine Receptor M1, Human |
115 |
0.88 |
Binding ≤ 1μM
|
|
ACM1_RAT |
P08482
|
Muscarinic Acetylcholine Receptor M1, Rat |
1000 |
0.76 |
Binding ≤ 1μM
|
|
ACM2_HUMAN |
P08172
|
Muscarinic Acetylcholine Receptor M2, Human |
25 |
0.97 |
Binding ≤ 1μM
|
|
ACM2_RAT |
P10980
|
Muscarinic Acetylcholine Receptor M2, Rat |
1000 |
0.76 |
Binding ≤ 1μM
|
|
ACM3_HUMAN |
P20309
|
Muscarinic Acetylcholine Receptor M3, Human |
100 |
0.89 |
Binding ≤ 1μM
|
|
ACM3_RAT |
P08483
|
Muscarinic Acetylcholine Receptor M3, Rat |
1000 |
0.76 |
Binding ≤ 1μM
|
|
ACM4_HUMAN |
P08173
|
Muscarinic Acetylcholine Receptor M4, Human |
130 |
0.88 |
Binding ≤ 1μM
|
|
ACM4_RAT |
P08485
|
Muscarinic Acetylcholine Receptor M4, Rat |
1000 |
0.76 |
Binding ≤ 1μM
|
|
ACM5_HUMAN |
P08912
|
Muscarinic Acetylcholine Receptor M5, Human |
600 |
0.79 |
Binding ≤ 1μM
|
|
ACH10_RAT |
Q9JLB5
|
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit, Rat |
224 |
0.85 |
Binding ≤ 1μM
|
|
ACHA3_RAT |
P04757
|
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit, Rat |
224 |
0.85 |
Binding ≤ 1μM
|
|
ACHA4_RAT |
P09483
|
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit, Rat |
224 |
0.85 |
Binding ≤ 1μM
|
|
Z104290 |
Z104290
|
Neuronal Acetylcholine Receptor; Alpha4/beta2 |
270 |
0.84 |
Binding ≤ 1μM
|
|
Z104303 |
Z104303
|
Muscarinic Acetylcholine Receptor |
1.9 |
1.11 |
Binding ≤ 10μM
|
|
ACM1_RAT |
P08482
|
Muscarinic Acetylcholine Receptor M1, Rat |
1000 |
0.76 |
Binding ≤ 10μM
|
|
ACM1_MOUSE |
P12657
|
Muscarinic Acetylcholine Receptor M1, Mouse |
3.33 |
1.08 |
Binding ≤ 10μM
|
|
ACM1_HUMAN |
P11229
|
Muscarinic Acetylcholine Receptor M1, Human |
115 |
0.88 |
Binding ≤ 10μM
|
|
ACM2_HUMAN |
P08172
|
Muscarinic Acetylcholine Receptor M2, Human |
1800 |
0.73 |
Binding ≤ 10μM
|
|
ACM2_RAT |
P10980
|
Muscarinic Acetylcholine Receptor M2, Rat |
1000 |
0.76 |
Binding ≤ 10μM
|
|
ACM3_RAT |
P08483
|
Muscarinic Acetylcholine Receptor M3, Rat |
1000 |
0.76 |
Binding ≤ 10μM
|
|
ACM3_HUMAN |
P20309
|
Muscarinic Acetylcholine Receptor M3, Human |
100 |
0.89 |
Binding ≤ 10μM
|
|
ACM4_RAT |
P08485
|
Muscarinic Acetylcholine Receptor M4, Rat |
1000 |
0.76 |
Binding ≤ 10μM
|
|
ACM4_HUMAN |
P08173
|
Muscarinic Acetylcholine Receptor M4, Human |
130 |
0.88 |
Binding ≤ 10μM
|
|
ACM5_HUMAN |
P08912
|
Muscarinic Acetylcholine Receptor M5, Human |
600 |
0.79 |
Binding ≤ 10μM
|
|
ACH10_RAT |
Q9JLB5
|
Neuronal Acetylcholine Receptor Protein Alpha-10 Subunit, Rat |
224 |
0.85 |
Binding ≤ 10μM
|
|
ACHA3_RAT |
P04757
|
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit, Rat |
224 |
0.85 |
Binding ≤ 10μM
|
|
ACHA4_RAT |
P09483
|
Neuronal Acetylcholine Receptor Protein Alpha-4 Subunit, Rat |
224 |
0.85 |
Binding ≤ 10μM
|
|
Z104290 |
Z104290
|
Neuronal Acetylcholine Receptor; Alpha4/beta2 |
270 |
0.84 |
Binding ≤ 10μM
|
|
Z80024 |
Z80024
|
A9 (Fibroblast Cells) |
1200 |
0.75 |
Functional ≤ 10μM
|
|
Z50512 |
Z50512
|
Cavia Porcellus |
150 |
0.87 |
Functional ≤ 10μM
|
|
ACM1_HUMAN |
P11229
|
Muscarinic Acetylcholine Receptor M1, Human |
545 |
0.80 |
Functional ≤ 10μM
|
|
ACM2_HUMAN |
P08172
|
Muscarinic Acetylcholine Receptor M2, Human |
10000 |
0.64 |
Functional ≤ 10μM
|
|
ACM3_HUMAN |
P20309
|
Muscarinic Acetylcholine Receptor M3, Human |
140 |
0.87 |
Functional ≤ 10μM
|
|
ACM4_HUMAN |
P08173
|
Muscarinic Acetylcholine Receptor M4, Human |
73 |
0.91 |
Functional ≤ 10μM
|
|
ACM5_HUMAN |
P08912
|
Muscarinic Acetylcholine Receptor M5, Human |
73 |
0.91 |
Functional ≤ 10μM
|
|
Z50592 |
Z50592
|
Oryctolagus Cuniculus |
545 |
0.80 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.99 |
4.36 |
-6.35 |
0 |
5 |
0 |
64 |
223.257 |
3 |
↓
|
|
|
|
Analogs
-
2384592
-
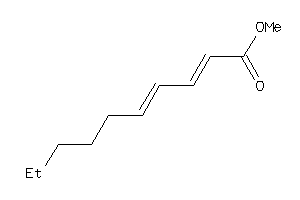
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.41 |
7.74 |
-5.09 |
0 |
2 |
0 |
26 |
180.247 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.47 |
-0.87 |
-2.95 |
1 |
2 |
0 |
29 |
104.149 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.76 |
7.03 |
-169.9 |
2 |
8 |
-3 |
156 |
314.229 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.08 |
7.04 |
-11.29 |
0 |
2 |
0 |
20 |
189.258 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.87 |
4.94 |
-8.03 |
0 |
6 |
0 |
74 |
237.211 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
-
19721454
-
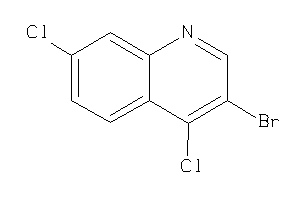
-
1020113
-
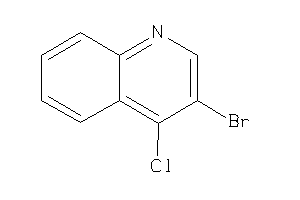
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.55 |
5.99 |
-4.39 |
0 |
1 |
0 |
13 |
242.503 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.55 |
5.99 |
-4.18 |
0 |
1 |
0 |
13 |
242.503 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.93 |
4.75 |
-5.44 |
0 |
2 |
0 |
22 |
238.084 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.66 |
-3.38 |
-43.17 |
1 |
6 |
-1 |
91 |
213.213 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.48 |
-0.82 |
-13.03 |
2 |
6 |
0 |
85 |
214.221 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.93 |
6.06 |
-6.29 |
0 |
4 |
0 |
47 |
227.304 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.93 |
6.06 |
-6.28 |
0 |
4 |
0 |
47 |
227.304 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.60 |
2.95 |
-8.37 |
3 |
4 |
0 |
68 |
154.169 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.94 |
1.01 |
-8.96 |
2 |
3 |
0 |
49 |
177.203 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.50 |
7.46 |
-5.84 |
0 |
3 |
0 |
43 |
269.094 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.45 |
7.24 |
-9.21 |
0 |
4 |
0 |
67 |
215.208 |
4 |
↓
|
|
|
|
Analogs
-
36040081
-
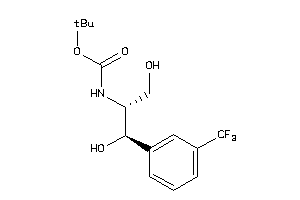
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.20 |
7.42 |
-9.25 |
2 |
4 |
0 |
59 |
421.337 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.60 |
6.92 |
-8.55 |
1 |
2 |
0 |
37 |
294.272 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.60 |
6.95 |
-8.71 |
1 |
2 |
0 |
37 |
294.272 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.17 |
12.92 |
-41.14 |
2 |
5 |
1 |
46 |
428.572 |
7 |
↓
|
|
|
|
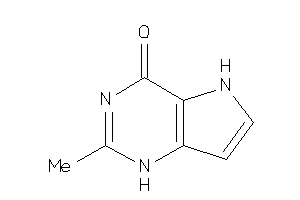
Analogs
-
39590671
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.00 |
0.53 |
-20.97 |
2 |
4 |
0 |
62 |
135.126 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.28 |
-0.91 |
-54.2 |
0 |
4 |
-1 |
61 |
134.118 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.46 |
0.03 |
-45.09 |
1 |
4 |
-1 |
65 |
134.118 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.17 |
3.67 |
-10.25 |
0 |
2 |
0 |
30 |
154.209 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.33 |
4.34 |
-6.46 |
1 |
4 |
0 |
55 |
210.62 |
2 |
↓
|
|
|
|
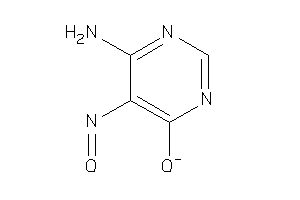
Analogs
-
25492098
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.30 |
-1.76 |
-11.94 |
5 |
5 |
0 |
98 |
126.119 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.65 |
0.12 |
-96.59 |
3 |
7 |
-2 |
135 |
169.096 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.25 |
1.46 |
-5.89 |
1 |
2 |
0 |
29 |
146.975 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.25 |
1.45 |
-3.25 |
1 |
2 |
0 |
29 |
146.975 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.58 |
5.15 |
-4.8 |
0 |
3 |
0 |
39 |
213.283 |
3 |
↓
|
|
|
|
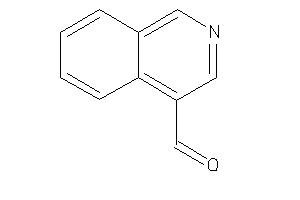
Analogs
-
2570382
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.72 |
2.89 |
-9.4 |
0 |
3 |
0 |
43 |
158.16 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.72 |
4.48 |
-11.68 |
0 |
2 |
0 |
26 |
144.177 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.56 |
5.46 |
-6.89 |
0 |
3 |
0 |
43 |
218.661 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.22 |
6.35 |
-3.72 |
0 |
2 |
0 |
26 |
204.678 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.80 |
5.88 |
-7.27 |
0 |
3 |
0 |
39 |
227.31 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.50 |
3.06 |
-41.65 |
0 |
8 |
-1 |
105 |
194.13 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.51 |
6.15 |
-7.06 |
0 |
2 |
0 |
26 |
190.242 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.99 |
1.46 |
-9.65 |
1 |
4 |
0 |
63 |
174.159 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.99 |
2.46 |
-46.68 |
0 |
4 |
-1 |
66 |
173.151 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.85 |
10.45 |
-14.76 |
2 |
8 |
0 |
111 |
464.518 |
11 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.85 |
13.2 |
-89.82 |
4 |
8 |
2 |
120 |
466.534 |
11 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.85 |
11.83 |
-49.45 |
3 |
8 |
1 |
115 |
465.526 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.67 |
4.49 |
-8.78 |
0 |
2 |
0 |
26 |
209.046 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.05 |
5.58 |
-51.49 |
0 |
3 |
-1 |
53 |
178.211 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.05 |
5.98 |
-30.02 |
1 |
3 |
0 |
54 |
179.219 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.47 |
4.44 |
-10.8 |
0 |
3 |
0 |
30 |
203.241 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KDM1A-1-E |
Lysine-specific Histone Demethylase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.24 |
Binding ≤ 10μM |
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KDM1A_HUMAN |
O60341
|
Lysine-specific Histone Demethylase 1, Human |
2000 |
0.24 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.04 |
10.08 |
-56.95 |
5 |
6 |
1 |
95 |
444.555 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.04 |
9.78 |
-21.4 |
4 |
6 |
0 |
93 |
443.547 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KDM1A-1-E |
Lysine-specific Histone Demethylase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.24 |
Binding ≤ 10μM |
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KDM1A_HUMAN |
O60341
|
Lysine-specific Histone Demethylase 1, Human |
2000 |
0.24 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.04 |
9.92 |
-60.61 |
5 |
6 |
1 |
95 |
444.555 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.04 |
9.6 |
-21.23 |
4 |
6 |
0 |
93 |
443.547 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KDM1A-1-E |
Lysine-specific Histone Demethylase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.24 |
Binding ≤ 10μM |
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KDM1A_HUMAN |
O60341
|
Lysine-specific Histone Demethylase 1, Human |
2000 |
0.24 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.04 |
9.92 |
-59.39 |
5 |
6 |
1 |
95 |
444.555 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.04 |
9.59 |
-20.43 |
4 |
6 |
0 |
93 |
443.547 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KDM1A-1-E |
Lysine-specific Histone Demethylase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2000 |
0.24 |
Binding ≤ 10μM |
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KDM1A_HUMAN |
O60341
|
Lysine-specific Histone Demethylase 1, Human |
2000 |
0.24 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.04 |
10.08 |
-59.05 |
5 |
6 |
1 |
95 |
444.555 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.04 |
9.79 |
-19.76 |
4 |
6 |
0 |
93 |
443.547 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.19 |
5.36 |
-7.32 |
0 |
4 |
0 |
39 |
209.245 |
4 |
↓
|
|
|
|
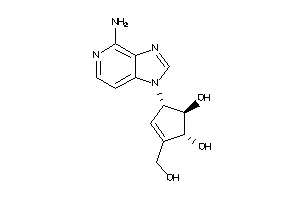
Analogs
-
29241818
-
-
29241824
-
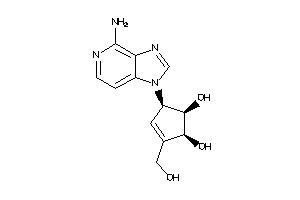
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.41 |
-1.22 |
-13.41 |
3 |
6 |
0 |
91 |
281.699 |
2 |
↓
|
|