|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.15 |
3.96 |
-8.85 |
3 |
6 |
0 |
96 |
462.627 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.15 |
3.64 |
-9.87 |
3 |
6 |
0 |
96 |
462.627 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.45 |
3.03 |
-49.18 |
1 |
5 |
1 |
49 |
334.436 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.53 |
5.74 |
-47.99 |
2 |
4 |
-1 |
81 |
325.469 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
94 |
0.55 |
Binding ≤ 10μM |
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES_HUMAN |
P22303
|
Acetylcholinesterase, Human |
150 |
0.53 |
Binding ≤ 1μM
|
|
ACES_TORCA |
P04058
|
Acetylcholinesterase, Torca |
11.4 |
0.62 |
Binding ≤ 1μM
|
|
ACES_ELEEL |
O42275
|
Acetylcholinesterase, Eleel |
120 |
0.54 |
Binding ≤ 1μM |
|
ACES_RAT |
P37136
|
Acetylcholinesterase, Rat |
114 |
0.54 |
Binding ≤ 1μM
|
|
ACES_BOVIN |
P23795
|
Acetylcholinesterase, Bovin |
23.6 |
0.59 |
Binding ≤ 1μM
|
|
ACES_RAT |
P37136
|
Acetylcholinesterase, Rat |
114 |
0.54 |
Binding ≤ 10μM
|
|
ACES_BOVIN |
P23795
|
Acetylcholinesterase, Bovin |
23.6 |
0.59 |
Binding ≤ 10μM
|
|
ACES_HUMAN |
P22303
|
Acetylcholinesterase, Human |
150 |
0.53 |
Binding ≤ 10μM
|
|
ACES_TORCA |
P04058
|
Acetylcholinesterase, Torca |
11.4 |
0.62 |
Binding ≤ 10μM
|
|
ACES_ELEEL |
O42275
|
Acetylcholinesterase, Eleel |
120 |
0.54 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.65 |
4.33 |
-56.88 |
4 |
3 |
1 |
61 |
243.33 |
0 |
↓
|
|
|
|
Analogs
-
15973598
-
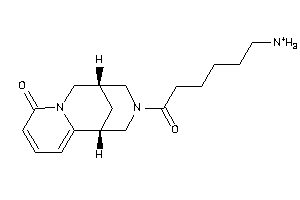
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.54 |
-2.71 |
-63.87 |
3 |
5 |
1 |
69 |
304.414 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
RARA-1-E |
Retinoic Acid Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Binding ≤ 10μM
|
|
RARB-2-E |
Retinoic Acid Receptor Beta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.52 |
Binding ≤ 10μM
|
|
RARG-2-E |
Retinoic Acid Receptor Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
17 |
0.49 |
Binding ≤ 10μM
|
|
RXRA-1-E |
Retinoid X Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
32 |
0.48 |
Binding ≤ 10μM
|
|
RXRB-2-E |
Retinoid X Receptor Beta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
15 |
0.50 |
Binding ≤ 10μM
|
|
RXRG-1-E |
Retinoid X Receptor Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.53 |
Binding ≤ 10μM
|
|
RARA-1-E |
Retinoic Acid Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
304 |
0.41 |
Functional ≤ 10μM
|
|
RARB-1-E |
Retinoic Acid Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
52 |
0.46 |
Functional ≤ 10μM
|
|
RARG-1-E |
Retinoic Acid Receptor Gamma (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
74 |
0.45 |
Functional ≤ 10μM
|
|
RXRA-1-E |
Retinoid X Receptor Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
13 |
0.50 |
Functional ≤ 10μM
|
|
RXRB-1-E |
Retinoid X Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.53 |
Functional ≤ 10μM
|
|
RXRG-2-E |
Retinoid X Receptor Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.53 |
Functional ≤ 10μM
|
|
Z50594-7-O |
Mus Musculus (cluster #7 Of 9), Other |
Other |
58 |
0.46 |
Functional ≤ 10μM
|
|
Z80156-5-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #5 Of 12), Other |
Other |
2 |
0.55 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
RARA_MOUSE |
P11416
|
Retinoic Acid Receptor Alpha, Mouse |
7 |
0.52 |
Binding ≤ 1μM
|
|
RARA_HUMAN |
P10276
|
Retinoic Acid Receptor Alpha, Human |
102 |
0.44 |
Binding ≤ 1μM
|
|
RARB_MOUSE |
P22605
|
Retinoic Acid Receptor Beta, Mouse |
7 |
0.52 |
Binding ≤ 1μM
|
|
RARB_HUMAN |
P10826
|
Retinoic Acid Receptor Beta, Human |
0.5 |
0.59 |
Binding ≤ 1μM
|
|
RARG_MOUSE |
P18911
|
Retinoic Acid Receptor Gamma, Mouse |
17 |
0.49 |
Binding ≤ 1μM
|
|
RARG_HUMAN |
P13631
|
Retinoic Acid Receptor Gamma, Human |
0.8 |
0.58 |
Binding ≤ 1μM
|
|
RXRA_MOUSE |
P28700
|
Retinoid X Receptor Alpha, Mouse |
200 |
0.43 |
Binding ≤ 1μM
|
|
RXRA_HUMAN |
P19793
|
Retinoid X Receptor Alpha, Human |
1.5 |
0.56 |
Binding ≤ 1μM
|
|
RXRB_MOUSE |
P28704
|
Retinoid X Receptor Beta, Mouse |
12 |
0.50 |
Binding ≤ 1μM
|
|
RXRB_HUMAN |
P28702
|
Retinoid X Receptor Beta, Human |
11 |
0.51 |
Binding ≤ 1μM
|
|
RXRG_MOUSE |
P28705
|
Retinoid X Receptor Gamma, Mouse |
124 |
0.44 |
Binding ≤ 1μM
|
|
RXRG_HUMAN |
P48443
|
Retinoid X Receptor Gamma, Human |
13 |
0.50 |
Binding ≤ 1μM
|
|
RARA_HUMAN |
P10276
|
Retinoic Acid Receptor Alpha, Human |
102 |
0.44 |
Binding ≤ 10μM
|
|
RARA_MOUSE |
P11416
|
Retinoic Acid Receptor Alpha, Mouse |
7 |
0.52 |
Binding ≤ 10μM
|
|
RARB_MOUSE |
P22605
|
Retinoic Acid Receptor Beta, Mouse |
7 |
0.52 |
Binding ≤ 10μM
|
|
RARB_HUMAN |
P10826
|
Retinoic Acid Receptor Beta, Human |
0.5 |
0.59 |
Binding ≤ 10μM
|
|
RARG_MOUSE |
P18911
|
Retinoic Acid Receptor Gamma, Mouse |
17 |
0.49 |
Binding ≤ 10μM
|
|
RARG_HUMAN |
P13631
|
Retinoic Acid Receptor Gamma, Human |
0.8 |
0.58 |
Binding ≤ 10μM
|
|
RXRA_MOUSE |
P28700
|
Retinoid X Receptor Alpha, Mouse |
200 |
0.43 |
Binding ≤ 10μM
|
|
RXRA_HUMAN |
P19793
|
Retinoid X Receptor Alpha, Human |
1.5 |
0.56 |
Binding ≤ 10μM
|
|
RXRB_MOUSE |
P28704
|
Retinoid X Receptor Beta, Mouse |
12 |
0.50 |
Binding ≤ 10μM
|
|
RXRB_HUMAN |
P28702
|
Retinoid X Receptor Beta, Human |
11 |
0.51 |
Binding ≤ 10μM
|
|
RXRG_HUMAN |
P48443
|
Retinoid X Receptor Gamma, Human |
13 |
0.50 |
Binding ≤ 10μM
|
|
RXRG_MOUSE |
P28705
|
Retinoid X Receptor Gamma, Mouse |
124 |
0.44 |
Binding ≤ 10μM
|
|
Z80156 |
Z80156
|
HL-60 (Promyeloblast Leukemia Cells) |
2 |
0.55 |
Functional ≤ 10μM
|
|
Z50594 |
Z50594
|
Mus Musculus |
58 |
0.46 |
Functional ≤ 10μM
|
|
RARA_HUMAN |
P10276
|
Retinoic Acid Receptor Alpha, Human |
102 |
0.44 |
Functional ≤ 10μM
|
|
RARB_HUMAN |
P10826
|
Retinoic Acid Receptor Beta, Human |
3 |
0.54 |
Functional ≤ 10μM
|
|
RARG_HUMAN |
P13631
|
Retinoic Acid Receptor Gamma, Human |
1 |
0.57 |
Functional ≤ 10μM
|
|
RXRA_RAT |
Q05343
|
Retinoid X Receptor Alpha, Rat |
13 |
0.50 |
Functional ≤ 10μM
|
|
RXRA_HUMAN |
P19793
|
Retinoid X Receptor Alpha, Human |
1.5 |
0.56 |
Functional ≤ 10μM
|
|
RXRB_HUMAN |
P28702
|
Retinoid X Receptor Beta, Human |
200 |
0.43 |
Functional ≤ 10μM
|
|
RXRG_HUMAN |
P48443
|
Retinoid X Receptor Gamma, Human |
11 |
0.51 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.80 |
11.58 |
-54.7 |
0 |
2 |
-1 |
40 |
299.434 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.80 |
11.01 |
-5.25 |
1 |
2 |
0 |
37 |
300.442 |
5 |
↓
|
|
|
|
Analogs
-
9034009
-
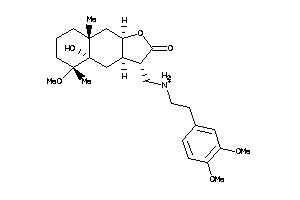
-
9034010
-
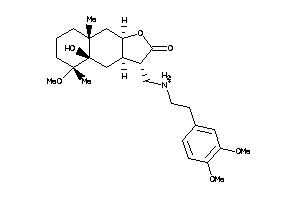
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3R,3aR,4aR,5R,8aR,9aR)-3-[[2-(3,4-dimethoxyphenyl)ethylamino]methyl]-4a-hydroxy-5-methoxy-5,8a-dime
(3R,3aR,4aR,5R,8aR,9aR)-3-[[2-(3…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.02 |
8.3 |
-56.63 |
3 |
7 |
1 |
91 |
462.607 |
8 |
↓
|
|
|
|
Analogs
-
9034009
-

-
9034010
-

Draw
Identity
99%
90%
80%
70%
Popular Name:
(3R,3aR,4aS,5R,8aR,9aR)-3-[[2-(3,4-dimethoxyphenyl)ethylamino]methyl]-4a-hydroxy-5-methoxy-5,8a-dime
(3R,3aR,4aS,5R,8aR,9aR)-3-[[2-(3…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.02 |
8.51 |
-53.54 |
3 |
7 |
1 |
91 |
462.607 |
8 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.20 |
-0.06 |
-8.36 |
1 |
4 |
0 |
59 |
274.297 |
2 |
↓
|
|
|
|
Analogs
-
5817054
-
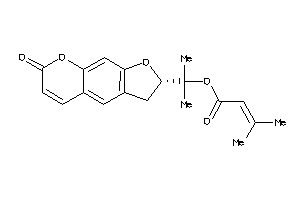
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.35 |
3.8 |
-15.31 |
0 |
5 |
0 |
65 |
328.364 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.58 |
-5.24 |
-19.6 |
2 |
7 |
0 |
85 |
317.345 |
5 |
↓
|
|
|
|
Analogs
-
1322496
-
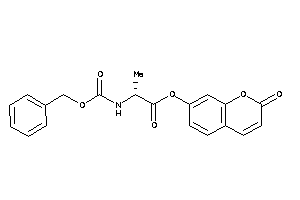
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.69 |
2.34 |
-15.4 |
1 |
7 |
0 |
94 |
367.357 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.33 |
-2.46 |
-9.66 |
2 |
6 |
0 |
72 |
334.379 |
5 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
32222023
-
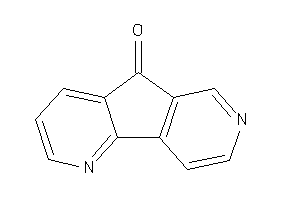
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.21 |
0.83 |
-9.8 |
0 |
2 |
0 |
29 |
257.292 |
1 |
↓
|
|
|
|
|
|
|
Analogs
-
5734580
-
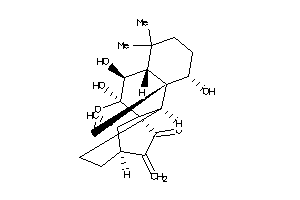
-
5741430
-
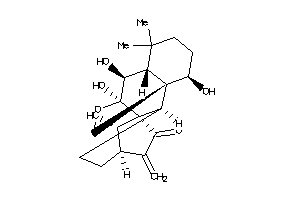
-
12661785
-
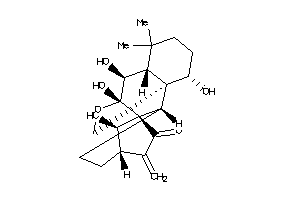
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1alpha,5beta,6beta,9beta,10alpha,14R)-1,6,7,14-tetrahydroxy-7,20-epoxykaur-16-en-15-one
(1alpha,5beta,6beta,9beta,10alph…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.33 |
-6.38 |
-11.98 |
4 |
6 |
0 |
107 |
364.438 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.92 |
3.18 |
-15.96 |
0 |
4 |
0 |
48 |
324.376 |
5 |
↓
|
|
|
|
Analogs
-
4027981
-
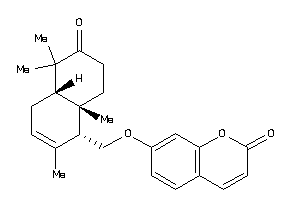
-
4028456
-
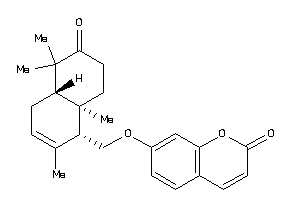
Draw
Identity
99%
90%
80%
70%
Popular Name:
7-[[(1S,4aS,8aR)-6-keto-2,5,5,8a-tetramethyl-4,4a,7,8-tetrahydro-1H-naphthalen-1-yl]methoxy]coumarin
7-[[(1S,4aS,8aR)-6-keto-2,5,5,8a…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.33 |
12.12 |
-15.56 |
0 |
4 |
0 |
57 |
380.484 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
3-[(3S,5S,8R,9S,10R,11R,13R,14S,17R)-5,11,14-trihydroxy-10,13-dimethyl-3-[(2R,3S,4R,5R,6S)-3,4,5-tri
3-[(3S,5S,8R,9S,10R,11R,13R,14S,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.09 |
-1.75 |
-18.8 |
6 |
10 |
0 |
166 |
552.661 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
3-[(3S,5S,8R,9S,10R,11R,13R,14S,17R)-5,11,14-trihydroxy-10,13-dimethyl-3-[(2R,3S,4R,5R,6R)-3,4,5-tri
3-[(3S,5S,8R,9S,10R,11R,13R,14S,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.09 |
-0.64 |
-17.77 |
6 |
10 |
0 |
166 |
552.661 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
3-[(3S,5S,8R,9S,10R,11R,13R,14S,17R)-5,11,14-trihydroxy-10,13-dimethyl-3-[(2R,3S,4S,5R,6S)-3,4,5-tri
3-[(3S,5S,8R,9S,10R,11R,13R,14S,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.09 |
-0.7 |
-17.1 |
6 |
10 |
0 |
166 |
552.661 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
3-[(3S,5S,8R,9S,10R,11R,13R,14S,17R)-5,11,14-trihydroxy-10,13-dimethyl-3-[(2R,3S,4S,5R,6R)-3,4,5-tri
3-[(3S,5S,8R,9S,10R,11R,13R,14S,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.09 |
0.03 |
-20.13 |
6 |
10 |
0 |
166 |
552.661 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(2S,3R,4S,5S,6S)-3,5-dihydroxy-2-[(1S,2R)-3-hydroxy-2-[(1S)-1-hydroxy-2-[(E)-(2-hydroxyphenyl)methy
[(2S,3R,4S,5S,6S)-3,5-dihydroxy-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.07 |
-4.57 |
-95.47 |
9 |
14 |
2 |
224 |
594.614 |
15 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.07 |
-6.38 |
-20.56 |
7 |
14 |
0 |
220 |
592.598 |
15 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.07 |
-5.44 |
-48.56 |
8 |
14 |
1 |
222 |
593.606 |
15 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(2S,3R,4R,5S,6S)-3,5-dihydroxy-2-[(1S,2R)-3-hydroxy-2-[(1S)-1-hydroxy-2-[(E)-(2-hydroxyphenyl)methy
[(2S,3R,4R,5S,6S)-3,5-dihydroxy-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.07 |
-3.96 |
-97 |
9 |
14 |
2 |
224 |
594.614 |
15 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.07 |
-5.68 |
-20.11 |
7 |
14 |
0 |
220 |
592.598 |
15 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.07 |
-4.82 |
-47.72 |
8 |
14 |
1 |
222 |
593.606 |
15 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(2S,3R,4S,5R,6S)-3,5-dihydroxy-2-[(1S,2R)-3-hydroxy-2-[(1S)-1-hydroxy-2-[(E)-(2-hydroxyphenyl)methy
[(2S,3R,4S,5R,6S)-3,5-dihydroxy-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.07 |
-3.75 |
-95.01 |
9 |
14 |
2 |
224 |
594.614 |
15 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.07 |
-5.56 |
-21.79 |
7 |
14 |
0 |
220 |
592.598 |
15 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.07 |
-4.6 |
-49.94 |
8 |
14 |
1 |
222 |
593.606 |
15 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(2S,3R,4R,5R,6S)-3,5-dihydroxy-2-[(1S,2R)-3-hydroxy-2-[(1S)-1-hydroxy-2-[(E)-(2-hydroxyphenyl)methy
[(2S,3R,4R,5R,6S)-3,5-dihydroxy-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.07 |
-2.88 |
-96.39 |
9 |
14 |
2 |
224 |
594.614 |
15 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.07 |
-4.61 |
-20.29 |
7 |
14 |
0 |
220 |
592.598 |
15 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.07 |
-3.75 |
-47.13 |
8 |
14 |
1 |
222 |
593.606 |
15 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.74 |
0.36 |
-47.15 |
2 |
7 |
1 |
73 |
357.434 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.74 |
0.85 |
-39.96 |
2 |
7 |
1 |
73 |
357.434 |
5 |
↓
|
|
|
|
|
|
|
Analogs
-
41719832
-
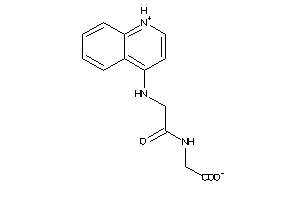
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.50 |
-3.7 |
-53.39 |
3 |
6 |
0 |
95 |
309.325 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.33 |
-0.02 |
-53.95 |
3 |
6 |
1 |
97 |
401.508 |
9 |
↓
|
|
|
|
Analogs
-
39289775
-
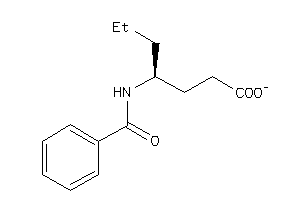
-
39289776
-
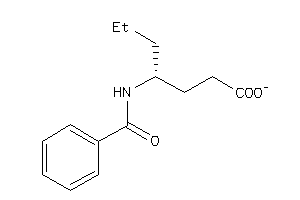
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.78 |
5.78 |
-47.86 |
1 |
4 |
-1 |
69 |
234.275 |
6 |
↓
|
|
|
|
Analogs
-
39289775
-

-
39289776
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.78 |
5.78 |
-47.85 |
1 |
4 |
-1 |
69 |
234.275 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.54 |
-0.86 |
-36.97 |
2 |
8 |
1 |
86 |
282.324 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.87 |
4.47 |
-46.93 |
1 |
10 |
-1 |
146 |
267.177 |
1 |
↓
|
|
|
|
Analogs
-
526070
-
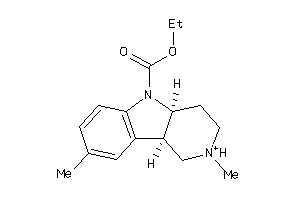
-
4565198
-
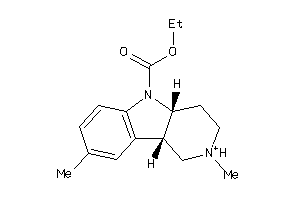
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4aR,9bR)-2,8-dimethyl-1,2,3,4,4a,9b-hexahydropyrido[4,3-b]indol-2-ium-5-carboxylic-acid-ethyl-ester
(4aR,9bR)-2,8-dimethyl-1,2,3,4,4…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.85 |
1.84 |
-43.98 |
1 |
4 |
1 |
33 |
275.372 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.91 |
-0.34 |
-62.15 |
2 |
7 |
-1 |
107 |
403.414 |
7 |
↓
|
|
|
|
Analogs
-
4029437
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.40 |
-0.89 |
-99.44 |
4 |
4 |
2 |
46 |
377.532 |
4 |
↓
|
|
|
|
Analogs
-
4029438
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.40 |
-0.89 |
-97.18 |
4 |
4 |
2 |
46 |
377.532 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3R,4S,5S,6R,7R,9R,11R,12R,13S,14R)-6-[(2R,3S,4R,6S)-4-(dimethylamino)-3-hydroxy-6-methyl-tetrahydro
(3R,4S,5S,6R,7R,9R,11R,12R,13S,1…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.90 |
6.01 |
-45.41 |
5 |
14 |
1 |
184 |
748.972 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.90 |
3.6 |
-17.6 |
4 |
14 |
0 |
183 |
747.964 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.62 |
10.94 |
-80.81 |
9 |
18 |
0 |
278 |
827.892 |
21 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.56 |
13.58 |
-37.05 |
4 |
13 |
1 |
155 |
811.997 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.08 |
11.92 |
-13.45 |
2 |
6 |
0 |
86 |
497.724 |
14 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.08 |
12.53 |
-31.72 |
3 |
6 |
1 |
87 |
498.732 |
14 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.08 |
11.79 |
-15.22 |
2 |
6 |
0 |
86 |
497.724 |
14 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.08 |
12.4 |
-32.9 |
3 |
6 |
1 |
87 |
498.732 |
14 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.08 |
12.53 |
-31.96 |
3 |
6 |
1 |
87 |
498.732 |
14 |
↓
|
|