|
|
Analogs
-
45286696
-
-
45484805
-
-
33505257
-
Draw
Identity
99%
90%
80%
70%
Popular Name:
[(1R,5aS,5bS,7aS,11aR,11bS,13aS,13bR)-1-acetoxy-5b,8,8,13a-tetramethyl-3-oxo-5,5a,6,7,7a,9,10,11,11b
[(1R,5aS,5bS,7aS,11aR,11bS,13aS,…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.40 |
14.75 |
-16.28 |
0 |
6 |
0 |
79 |
486.649 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.09 |
6.85 |
-13.58 |
1 |
7 |
0 |
99 |
374.389 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.09 |
7.39 |
-51.44 |
0 |
7 |
-1 |
102 |
373.381 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.09 |
7.12 |
-13.02 |
1 |
7 |
0 |
99 |
374.389 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.09 |
7.66 |
-50.23 |
0 |
7 |
-1 |
102 |
373.381 |
4 |
↓
|
|
|
|
Analogs
-
4098165
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.12 |
10.49 |
-8.63 |
0 |
6 |
0 |
71 |
374.433 |
5 |
↓
|
|
|
|
Analogs
-
4098165
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.12 |
9.32 |
-10.54 |
0 |
6 |
0 |
71 |
374.433 |
5 |
↓
|
|
|
|
Analogs
-
4098165
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.12 |
10.61 |
-8.28 |
0 |
6 |
0 |
71 |
374.433 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
O1-[(3aR,5R,5aS,8aR,9S,9aS)-5,8a-dimethyl-1-methylene-2,8-dioxo-3a,4,5,5a,9,9a-hexahydroazuleno[6,5-
O1-[(3aR,5R,5aS,8aR,9S,9aS)-5,8a…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.79 |
17.44 |
-34.81 |
0 |
10 |
0 |
139 |
620.695 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
O1-[(3aR,5S,5aS,8aS,9R,9aS)-5,8a-dimethyl-1-methylene-2,8-dioxo-3a,4,5,5a,9,9a-hexahydroazuleno[6,5-
O1-[(3aR,5S,5aS,8aS,9R,9aS)-5,8a…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.79 |
17.35 |
-37.38 |
0 |
10 |
0 |
139 |
620.695 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
O1-[(3aS,5R,5aS,8aR,9S,9aS)-5,8a-dimethyl-1-methylene-2,8-dioxo-3a,4,5,5a,9,9a-hexahydroazuleno[6,5-
O1-[(3aS,5R,5aS,8aR,9S,9aS)-5,8a…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.79 |
17.58 |
-33.99 |
0 |
10 |
0 |
139 |
620.695 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
O1-[(3aR,5S,5aS,8aS,9R,9aS)-5,8a-dimethyl-1-methylene-2,8-dioxo-3a,4,5,5a,9,9a-hexahydroazuleno[6,5-
O1-[(3aR,5S,5aS,8aS,9R,9aS)-5,8a…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.79 |
17.7 |
-33.34 |
0 |
10 |
0 |
139 |
620.695 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aR,5R,5aS,8aS,9S,9aR)-9-hydroxy-5,8a-trimethyl-methylene-spiro[3a,4,5,5a,9,9a-hexahydroazuleno[
(1R,3aR,5R,5aS,8aS,9S,9aR)-9-hyd…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.49 |
9.67 |
-15.87 |
1 |
7 |
0 |
107 |
494.584 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aR,5S,5aS,8aS,9S,9aR)-9-hydroxy-5,8a-trimethyl-methylene-spiro[3a,4,5,5a,9,9a-hexahydroazuleno[
(1R,3aR,5S,5aS,8aS,9S,9aR)-9-hyd…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.49 |
9.64 |
-15.79 |
1 |
7 |
0 |
107 |
494.584 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aS,5R,5aS,8aS,9S,9aR)-9-hydroxy-5,8a-trimethyl-methylene-spiro[3a,4,5,5a,9,9a-hexahydroazuleno[
(1R,3aS,5R,5aS,8aS,9S,9aR)-9-hyd…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.49 |
10.59 |
-16.96 |
1 |
7 |
0 |
107 |
494.584 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(1R,3aS,5S,5aS,8aS,9S,9aR)-9-hydroxy-5,8a-trimethyl-methylene-spiro[3a,4,5,5a,9,9a-hexahydroazuleno[
(1R,3aS,5S,5aS,8aS,9S,9aR)-9-hyd…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.49 |
10.58 |
-16.77 |
1 |
7 |
0 |
107 |
494.584 |
0 |
↓
|
|
|
|
Analogs
-
4492886
-
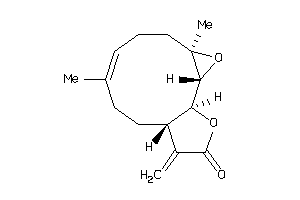
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.09 |
7.25 |
-11.58 |
0 |
3 |
0 |
39 |
248.322 |
0 |
↓
|
|
|
|
Analogs
-
12402558
-
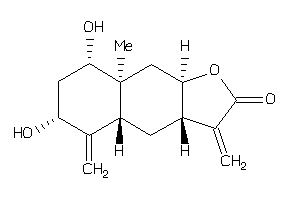
-
12402559
-
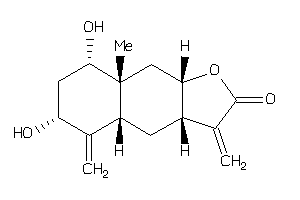
-
12402560
-
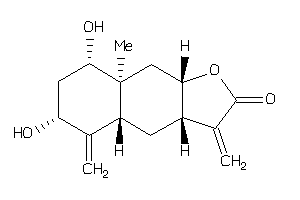
-
15056082
-
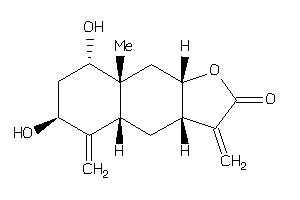
-
15056084
-
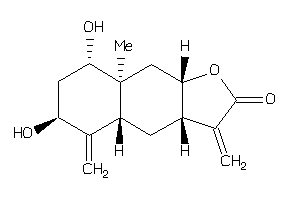
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3aS,4aR,6R,8S,8aR,9aR)-6,8-dihydroxy-8a-methyl-3,5-dimethylene-3a,4,4a,6,7,8,9,9a-octahydrobenzo[f]
(3aS,4aR,6R,8S,8aR,9aR)-6,8-dihy…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.39 |
-0.19 |
-9.36 |
2 |
4 |
0 |
67 |
264.321 |
0 |
↓
|
|
|
|
Analogs
-
12402559
-

-
12402560
-

-
15056082
-

-
15056084
-

-
15056087
-
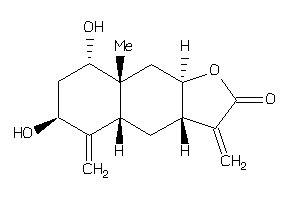
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3aS,4aR,6R,8S,8aS,9aR)-6,8-dihydroxy-8a-methyl-3,5-dimethylene-3a,4,4a,6,7,8,9,9a-octahydrobenzo[f]
(3aS,4aR,6R,8S,8aS,9aR)-6,8-dihy…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.39 |
-2.42 |
-11.29 |
2 |
4 |
0 |
67 |
264.321 |
0 |
↓
|
|
|
|
Analogs
-
12402560
-

-
15056082
-

-
15056084
-

-
15056087
-

-
15056089
-
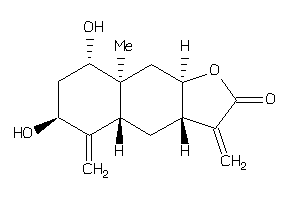
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3aS,4aR,6R,8S,8aR,9aS)-6,8-dihydroxy-8a-methyl-3,5-dimethylene-3a,4,4a,6,7,8,9,9a-octahydrobenzo[f]
(3aS,4aR,6R,8S,8aR,9aS)-6,8-dihy…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.39 |
-2.03 |
-12.63 |
2 |
4 |
0 |
67 |
264.321 |
0 |
↓
|
|
|
|
Analogs
-
15056082
-

-
15056084
-

-
15056087
-

-
15056089
-

-
4098119
-
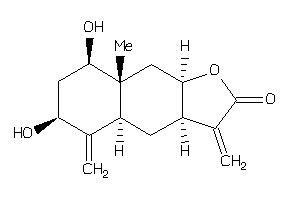
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3aS,4aR,6R,8S,8aS,9aS)-6,8-dihydroxy-8a-methyl-3,5-dimethylene-3a,4,4a,6,7,8,9,9a-octahydrobenzo[f]
(3aS,4aR,6R,8S,8aS,9aS)-6,8-dihy…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.39 |
-2.08 |
-14.33 |
2 |
4 |
0 |
67 |
264.321 |
0 |
↓
|
|
|
|
|
|
|
Analogs
-
12402928
-
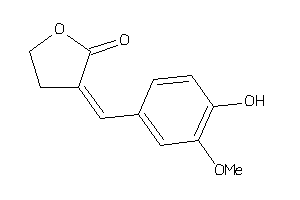
-
1591951
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.09 |
2.18 |
-13.47 |
0 |
4 |
0 |
45 |
234.251 |
3 |
↓
|
|
|
|
Analogs
-
1628265
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.26 |
1.14 |
-16.52 |
1 |
4 |
0 |
64 |
232.235 |
2 |
↓
|
|
|
|
Analogs
-
1646550
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.08 |
0.43 |
-16.93 |
1 |
4 |
0 |
64 |
232.235 |
2 |
↓
|
|
|
|
Analogs
-
1715295
-
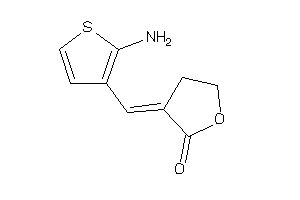
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.46 |
-0.97 |
-11.26 |
2 |
3 |
0 |
52 |
195.243 |
1 |
↓
|
|
|
|
Analogs
-
34553088
-
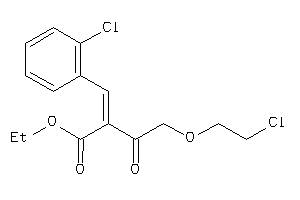
-
1747666
-
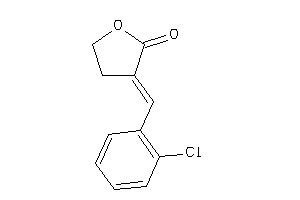
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.90 |
1.56 |
-8.08 |
0 |
2 |
0 |
26 |
208.644 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.24 |
3.98 |
-21.43 |
4 |
9 |
0 |
136 |
389.437 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.24 |
3.73 |
-60.3 |
3 |
9 |
-1 |
138 |
388.429 |
6 |
↓
|
|
|
|
Analogs
-
8649676
-
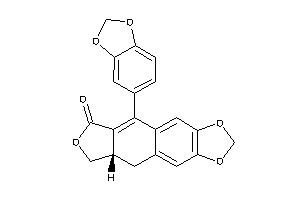
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.15 |
8.29 |
-15.67 |
0 |
6 |
0 |
63 |
366.369 |
3 |
↓
|
|
|
|
Analogs
-
33704877
-
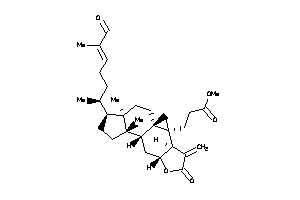
-
33704879
-
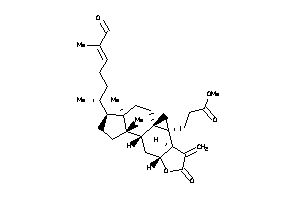
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.94 |
14.08 |
-53.73 |
0 |
5 |
-1 |
84 |
481.653 |
8 |
↓
|
|
|
|
Analogs
-
33704877
-

-
33704879
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.94 |
14.12 |
-50.8 |
0 |
5 |
-1 |
84 |
481.653 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.01 |
8.94 |
-8.77 |
0 |
2 |
0 |
26 |
234.339 |
1 |
↓
|
|
|
|
Analogs
-
32139522
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.04 |
16.02 |
-10.76 |
0 |
4 |
0 |
53 |
482.705 |
8 |
↓
|
|
|
|
Analogs
-
32139522
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.04 |
16 |
-9.24 |
0 |
4 |
0 |
53 |
482.705 |
8 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.05 |
1.48 |
-13.17 |
3 |
5 |
0 |
87 |
350.455 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.22 |
3.81 |
-8.73 |
0 |
2 |
0 |
26 |
112.128 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
|
KPCD-2-E |
Protein Kinase C Delta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1890 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.33 |
11.61 |
-12.95 |
1 |
4 |
0 |
56 |
368.558 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
|
KPCD-2-E |
Protein Kinase C Delta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1890 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.33 |
11.57 |
-13.2 |
1 |
4 |
0 |
56 |
368.558 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
|
KPCA-2-E |
Protein Kinase C Alpha (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
5874 |
0.28 |
Binding ≤ 10μM
|
|
KPCD-2-E |
Protein Kinase C Delta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2140 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.33 |
11.51 |
-12.34 |
1 |
4 |
0 |
56 |
368.558 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
|
KPCA-2-E |
Protein Kinase C Alpha (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
5874 |
0.28 |
Binding ≤ 10μM
|
|
KPCD-2-E |
Protein Kinase C Delta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
2140 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.33 |
11.33 |
-12.82 |
1 |
4 |
0 |
56 |
368.558 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
|
KPCA-2-E |
Protein Kinase C Alpha (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
113 |
0.36 |
Binding ≤ 10μM
|
|
KPCD-2-E |
Protein Kinase C Delta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
606 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.75 |
12.2 |
-13.94 |
1 |
4 |
0 |
56 |
398.609 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
|
KPCA-2-E |
Protein Kinase C Alpha (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
113 |
0.36 |
Binding ≤ 10μM
|
|
KPCD-2-E |
Protein Kinase C Delta (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
606 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.75 |
12.2 |
-13.8 |
1 |
4 |
0 |
56 |
398.609 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.13 |
5.78 |
-69.58 |
3 |
6 |
1 |
80 |
410.49 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.13 |
4.39 |
-18.21 |
2 |
6 |
0 |
79 |
409.482 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.13 |
6.03 |
-56.06 |
3 |
6 |
1 |
80 |
410.49 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.13 |
4.44 |
-17.47 |
2 |
6 |
0 |
79 |
409.482 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
|
OPRD_RAT |
P33533
|
Delta Opioid Receptor, Rat |
12.5 |
0.37 |
Binding ≤ 1μM
|
|
OPRD_MOUSE |
P32300
|
Delta Opioid Receptor, Mouse |
12.5 |
0.37 |
Binding ≤ 1μM
|
|
OPRD_HUMAN |
P41143
|
Delta Opioid Receptor, Human |
6 |
0.38 |
Binding ≤ 1μM
|
|
OPRK_RAT |
P34975
|
Kappa Opioid Receptor, Rat |
12.5 |
0.37 |
Binding ≤ 1μM
|
|
OPRK_MOUSE |
P33534
|
Kappa Opioid Receptor, Mouse |
12.5 |
0.37 |
Binding ≤ 1μM
|
|
OPRK_HUMAN |
P41145
|
Kappa Opioid Receptor, Human |
6 |
0.38 |
Binding ≤ 1μM
|
|
OPRM_HUMAN |
P35372
|
Mu Opioid Receptor, Human |
6 |
0.38 |
Binding ≤ 1μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
12.5 |
0.37 |
Binding ≤ 1μM
|
|
OPRM_BOVIN |
P79350
|
Mu Opioid Receptor, Bovin |
1.8 |
0.41 |
Binding ≤ 1μM
|
|
OPRM_MOUSE |
P42866
|
Mu Opioid Receptor, Mouse |
12.5 |
0.37 |
Binding ≤ 1μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
12.5 |
0.37 |
Binding ≤ 1μM
|
|
SGMR1_MOUSE |
O55242
|
Sigma Opioid Receptor, Mouse |
12.5 |
0.37 |
Binding ≤ 1μM
|
|
OPRD_HUMAN |
P41143
|
Delta Opioid Receptor, Human |
6 |
0.38 |
Binding ≤ 10μM
|
|
OPRD_RAT |
P33533
|
Delta Opioid Receptor, Rat |
12.5 |
0.37 |
Binding ≤ 10μM
|
|
OPRD_MOUSE |
P32300
|
Delta Opioid Receptor, Mouse |
12.5 |
0.37 |
Binding ≤ 10μM
|
|
OPRK_HUMAN |
P41145
|
Kappa Opioid Receptor, Human |
6 |
0.38 |
Binding ≤ 10μM
|
|
OPRK_RAT |
P34975
|
Kappa Opioid Receptor, Rat |
12.5 |
0.37 |
Binding ≤ 10μM
|
|
OPRK_MOUSE |
P33534
|
Kappa Opioid Receptor, Mouse |
12.5 |
0.37 |
Binding ≤ 10μM
|
|
OPRM_HUMAN |
P35372
|
Mu Opioid Receptor, Human |
6 |
0.38 |
Binding ≤ 10μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
12.5 |
0.37 |
Binding ≤ 10μM
|
|
OPRM_BOVIN |
P79350
|
Mu Opioid Receptor, Bovin |
1.8 |
0.41 |
Binding ≤ 10μM
|
|
OPRM_MOUSE |
P42866
|
Mu Opioid Receptor, Mouse |
12.5 |
0.37 |
Binding ≤ 10μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
12.5 |
0.37 |
Binding ≤ 10μM
|
|
SGMR1_MOUSE |
O55242
|
Sigma Opioid Receptor, Mouse |
12.5 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.13 |
6.09 |
-66.48 |
3 |
6 |
1 |
80 |
410.49 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.13 |
4.25 |
-19.2 |
2 |
6 |
0 |
79 |
409.482 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
|
OPRD_RAT |
P33533
|
Delta Opioid Receptor, Rat |
12.5 |
0.37 |
Binding ≤ 1μM
|
|
OPRD_MOUSE |
P32300
|
Delta Opioid Receptor, Mouse |
12.5 |
0.37 |
Binding ≤ 1μM
|
|
OPRD_HUMAN |
P41143
|
Delta Opioid Receptor, Human |
6 |
0.38 |
Binding ≤ 1μM
|
|
OPRK_RAT |
P34975
|
Kappa Opioid Receptor, Rat |
12.5 |
0.37 |
Binding ≤ 1μM
|
|
OPRK_MOUSE |
P33534
|
Kappa Opioid Receptor, Mouse |
12.5 |
0.37 |
Binding ≤ 1μM
|
|
OPRK_HUMAN |
P41145
|
Kappa Opioid Receptor, Human |
6 |
0.38 |
Binding ≤ 1μM
|
|
OPRM_HUMAN |
P35372
|
Mu Opioid Receptor, Human |
6 |
0.38 |
Binding ≤ 1μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
12.5 |
0.37 |
Binding ≤ 1μM
|
|
OPRM_BOVIN |
P79350
|
Mu Opioid Receptor, Bovin |
1.8 |
0.41 |
Binding ≤ 1μM
|
|
OPRM_MOUSE |
P42866
|
Mu Opioid Receptor, Mouse |
12.5 |
0.37 |
Binding ≤ 1μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
12.5 |
0.37 |
Binding ≤ 1μM
|
|
SGMR1_MOUSE |
O55242
|
Sigma Opioid Receptor, Mouse |
12.5 |
0.37 |
Binding ≤ 1μM
|
|
OPRD_HUMAN |
P41143
|
Delta Opioid Receptor, Human |
6 |
0.38 |
Binding ≤ 10μM
|
|
OPRD_RAT |
P33533
|
Delta Opioid Receptor, Rat |
12.5 |
0.37 |
Binding ≤ 10μM
|
|
OPRD_MOUSE |
P32300
|
Delta Opioid Receptor, Mouse |
12.5 |
0.37 |
Binding ≤ 10μM
|
|
OPRK_HUMAN |
P41145
|
Kappa Opioid Receptor, Human |
6 |
0.38 |
Binding ≤ 10μM
|
|
OPRK_RAT |
P34975
|
Kappa Opioid Receptor, Rat |
12.5 |
0.37 |
Binding ≤ 10μM
|
|
OPRK_MOUSE |
P33534
|
Kappa Opioid Receptor, Mouse |
12.5 |
0.37 |
Binding ≤ 10μM
|
|
OPRM_HUMAN |
P35372
|
Mu Opioid Receptor, Human |
6 |
0.38 |
Binding ≤ 10μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
12.5 |
0.37 |
Binding ≤ 10μM
|
|
OPRM_BOVIN |
P79350
|
Mu Opioid Receptor, Bovin |
1.8 |
0.41 |
Binding ≤ 10μM
|
|
OPRM_MOUSE |
P42866
|
Mu Opioid Receptor, Mouse |
12.5 |
0.37 |
Binding ≤ 10μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
12.5 |
0.37 |
Binding ≤ 10μM
|
|
SGMR1_MOUSE |
O55242
|
Sigma Opioid Receptor, Mouse |
12.5 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.13 |
6.58 |
-50.21 |
3 |
6 |
1 |
80 |
410.49 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.13 |
4.79 |
-16.13 |
2 |
6 |
0 |
79 |
409.482 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.17 |
5.12 |
-59.43 |
3 |
6 |
1 |
80 |
396.463 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.17 |
3.57 |
-17.68 |
2 |
6 |
0 |
79 |
395.455 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.17 |
4.88 |
-68.75 |
3 |
6 |
1 |
80 |
396.463 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.17 |
3.54 |
-17.69 |
2 |
6 |
0 |
79 |
395.455 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.17 |
6.45 |
-54.66 |
3 |
6 |
1 |
80 |
396.463 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.17 |
4.74 |
-19.4 |
2 |
6 |
0 |
79 |
395.455 |
2 |
↓
|
|