|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNN1-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.31 |
Binding ≤ 10μM
|
|
KCNN3-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.31 |
Binding ≤ 10μM
|
|
KCNN1-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.31 |
Functional ≤ 10μM
|
|
KCNN2-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.31 |
Functional ≤ 10μM
|
|
KCNN3-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.31 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
3 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.52 |
16.38 |
-67.76 |
2 |
4 |
2 |
32 |
494.642 |
0 |
↓
|
|
|
|
Analogs
-
2583341
-
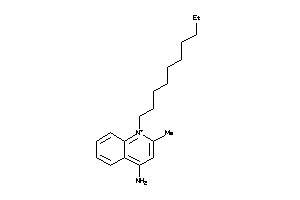
-
5820628
-
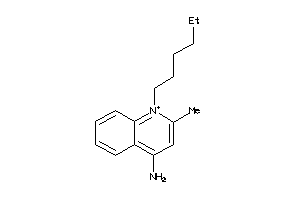
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNN1-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1700 |
0.24 |
Binding ≤ 10μM
|
|
KCNN2-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1700 |
0.24 |
Binding ≤ 10μM
|
|
KCNN3-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.23 |
Binding ≤ 10μM
|
|
KCNN1-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
900 |
0.25 |
Functional ≤ 10μM
|
|
KCNN2-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
900 |
0.25 |
Functional ≤ 10μM
|
|
KCNN3-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
900 |
0.25 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
316 |
0.27 |
Functional ≤ 10μM
|
|
Z50597-3-O |
Rattus Norvegicus (cluster #3 Of 12), Other |
Other |
900 |
0.25 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.11 |
15.83 |
-56.47 |
4 |
4 |
2 |
60 |
456.678 |
11 |
↓
|
|
|
|
Analogs
-
4245635
-
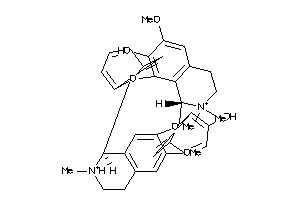
-
38153422
-
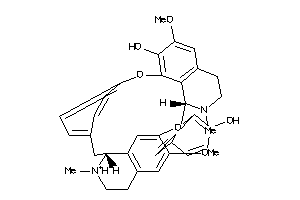
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA7-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
4790 |
0.17 |
Binding ≤ 10μM
|
|
KCNN1-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.17 |
Binding ≤ 10μM
|
|
KCNN2-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.17 |
Binding ≤ 10μM
|
|
KCNN3-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.17 |
Binding ≤ 10μM
|
|
KCNN1-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7500 |
0.16 |
Functional ≤ 10μM
|
|
KCNN2-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7500 |
0.16 |
Functional ≤ 10μM
|
|
KCNN3-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7500 |
0.16 |
Functional ≤ 10μM
|
|
Z104283-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #1 Of 4), Other |
Other |
9720 |
0.16 |
Binding ≤ 10μM
|
|
Z104288-1-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #1 Of 2), Other |
Other |
2000 |
0.18 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.99 |
3.03 |
-94.42 |
3 |
8 |
2 |
81 |
610.751 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNN1-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.39 |
Binding ≤ 10μM
|
|
KCNN2-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
KCNN3-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
KCNN1-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.38 |
Functional ≤ 10μM
|
|
KCNN2-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.38 |
Functional ≤ 10μM
|
|
KCNN3-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.38 |
Functional ≤ 10μM
|
|
Z50597-3-O |
Rattus Norvegicus (cluster #3 Of 12), Other |
Other |
3 |
0.37 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.14 |
13.71 |
-72.94 |
2 |
4 |
2 |
32 |
426.608 |
0 |
↓
|
|
|
|
Analogs
-
53158792
-
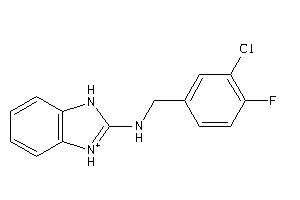
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNN3-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
200 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.29 |
-2.17 |
-9.97 |
2 |
3 |
0 |
40 |
292.169 |
3 |
↓
|
|
|
|
Analogs
-
53158792
-

Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNN3-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
640 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.68 |
-2.04 |
-8.92 |
2 |
3 |
0 |
40 |
257.724 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 28 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNN3-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2700 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.01 |
8.85 |
-27.47 |
3 |
3 |
1 |
42 |
224.287 |
3 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNN3-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
130 |
0.48 |
Binding ≤ 10μM
|
|
KCNN3-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.97 |
9.96 |
-26.48 |
3 |
3 |
1 |
42 |
264.352 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNN3-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
130 |
0.48 |
Binding ≤ 10μM
|
|
KCNN3-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
450 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.97 |
9.83 |
-30.06 |
3 |
3 |
1 |
42 |
264.352 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNN3-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
540 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.60 |
9.93 |
-27.38 |
3 |
3 |
1 |
42 |
270.381 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNN3-1-E |
Small Conductance Calcium-activated Potassium Channel Protein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
540 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.60 |
9.16 |
-26.99 |
3 |
3 |
1 |
42 |
270.381 |
2 |
↓
|
|
|
|
|
|
|
|