|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 51 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-3-E |
Prothrombin (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
150 |
1.06 |
Binding ≤ 10μM
|
|
TRY1-2-E |
Trypsin I (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
360 |
1.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.33 |
3.81 |
-27.95 |
4 |
3 |
1 |
55 |
128.199 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
C1R-1-E |
Complement C1r (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1400 |
0.46 |
Binding ≤ 10μM |
|
C1S-1-E |
Complement C1s (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1250 |
0.46 |
Binding ≤ 10μM |
|
KLK1-1-E |
Kallikrein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9890 |
0.39 |
Binding ≤ 10μM |
|
PLMN-1-E |
Plasminogen (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6680 |
0.40 |
Binding ≤ 10μM |
|
THRB-4-E |
Prothrombin (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
510 |
0.49 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.24 |
1.15 |
-8.83 |
0 |
3 |
0 |
43 |
349.127 |
1 |
↓
|
|
|
|
Analogs
-
4426033
-
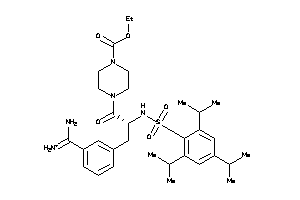
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.30 |
10.74 |
-41.24 |
5 |
10 |
1 |
148 |
614.833 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4 |
0.37 |
Binding ≤ 10μM
|
|
FA9-1-E |
Coagulation Factor IX (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4500 |
0.23 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1200 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.11 |
5.22 |
-21.63 |
2 |
8 |
0 |
107 |
479.986 |
5 |
↓
|
|
Ref
Reference (pH 7)
|
1.18 |
4.09 |
-51.74 |
3 |
8 |
1 |
112 |
480.994 |
5 |
↓
|
|
|
|
Analogs
-
3917722
-
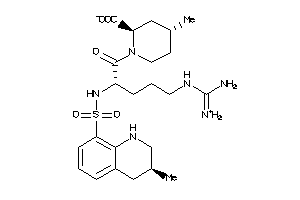
-
4096592
-
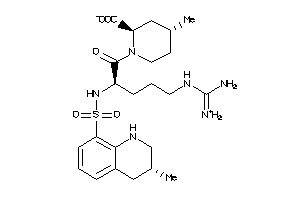
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1 |
0.36 |
Binding ≤ 10μM
|
|
TRY1-3-E |
Trypsin I (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
5000 |
0.21 |
Binding ≤ 10μM |
|
THRB-1-E |
Thrombin (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
38 |
0.30 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.78 |
6.76 |
-106.51 |
7 |
11 |
0 |
182 |
508.645 |
10 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
26 |
0.30 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.50 |
7.05 |
-77.8 |
6 |
10 |
0 |
154 |
477.569 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.50 |
7.51 |
-87.34 |
7 |
10 |
1 |
155 |
478.577 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.50 |
8.32 |
-110.06 |
8 |
10 |
2 |
160 |
479.585 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.72 |
3.62 |
-47.9 |
7 |
9 |
1 |
167 |
462.511 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.03 |
4.68 |
-44.4 |
7 |
9 |
1 |
159 |
475.554 |
6 |
↓
|
|
|
|
Analogs
-
4245610
-
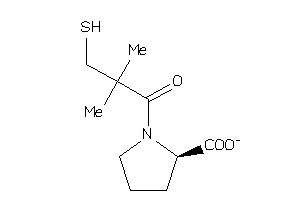
-
32218440
-
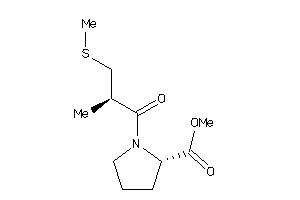
-
32218441
-
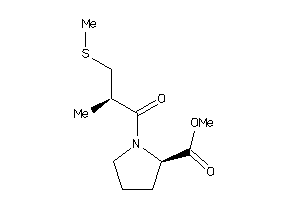
-
32218442
-
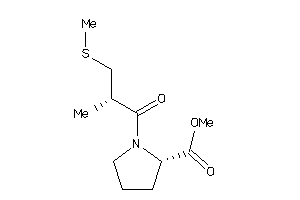
-
32218443
-
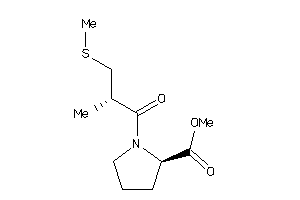
Draw
Identity
99%
90%
80%
70%
Vendors
And 37 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACE-1-E |
Angiotensin-converting Enzyme (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
23 |
0.76 |
Binding ≤ 10μM
|
|
ACE-1-E |
Angiotensin-converting Enzyme (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
70 |
0.72 |
Binding ≤ 10μM
|
|
ACE2-1-E |
Angiotensin-converting Enzyme 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
9 |
0.80 |
Binding ≤ 10μM
|
|
AMPB-1-E |
Aminopeptidase B (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.57 |
Binding ≤ 10μM |
|
AMPE-1-E |
Aminopeptidase A (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.57 |
Binding ≤ 10μM |
|
AMPN-1-E |
Aminopeptidase N (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
2000 |
0.57 |
Binding ≤ 10μM |
|
LKHA4-2-E |
Leukotriene A4 Hydrolase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
70 |
0.72 |
Binding ≤ 10μM |
|
RENI-1-E |
Renin (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.87 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
48 |
0.73 |
Binding ≤ 10μM
|
|
DAPE-1-B |
Succinyl-diaminopimelate Desuccinylase (cluster #1 Of 1), Bacterial |
Bacteria |
3300 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.09 |
5.14 |
-56.41 |
0 |
4 |
-1 |
60 |
216.282 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.45 |
-1.53 |
-64.17 |
4 |
7 |
1 |
108 |
533.527 |
7 |
↓
|
|
|
|
Analogs
-
34025326
-
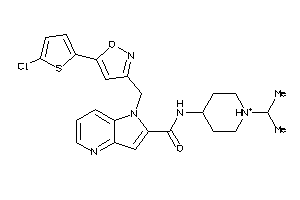
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.36 |
Binding ≤ 10μM
|
|
THRB-4-E |
Prothrombin (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
2760 |
0.24 |
Binding ≤ 10μM
|
|
THRB-1-E |
Thrombin (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
420 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.58 |
13.43 |
-41.46 |
2 |
6 |
1 |
65 |
484.045 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1 |
0.42 |
Binding ≤ 10μM
|
|
TRY1-1-E |
Trypsin I (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1800 |
0.27 |
Binding ≤ 10μM
|
|
TRY2-1-E |
Trypsin II (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1800 |
0.27 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.23 |
7.32 |
-45.79 |
5 |
8 |
1 |
116 |
407.498 |
8 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.23 |
6.88 |
-17.19 |
4 |
8 |
0 |
115 |
406.49 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.23 |
7.69 |
-79.08 |
6 |
8 |
2 |
117 |
408.506 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.40 |
-6.86 |
-92.71 |
6 |
10 |
1 |
167 |
538.65 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 85 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-2-E |
Coagulation Factor X (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
410 |
0.99 |
Binding ≤ 10μM
|
|
THRB-5-E |
Prothrombin (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
300 |
1.01 |
Binding ≤ 10μM
|
|
TRY1-1-E |
Trypsin I (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
31 |
1.17 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.32 |
2.36 |
-30.28 |
4 |
2 |
1 |
52 |
121.163 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3100 |
0.23 |
Binding ≤ 10μM
|
|
Z50592-3-O |
Oryctolagus Cuniculus (cluster #3 Of 8), Other |
Other |
329 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.78 |
-1.89 |
-17.12 |
2 |
9 |
0 |
111 |
459.506 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ELNE-3-E |
Neutrophil Elastase (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.77 |
Binding ≤ 10μM
|
|
KLKB1-1-E |
Plasma Kallikrein (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6600 |
0.48 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
6900 |
0.48 |
Binding ≤ 10μM
|
|
UROK-1-E |
Urokinase-type Plasminogen Activator (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2500 |
0.52 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.63 |
2.54 |
-5.1 |
0 |
3 |
0 |
35 |
224.622 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 3 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
8800 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
2.29 |
-7.63 |
0 |
6 |
0 |
80 |
314.128 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 6 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ELNE-1-E |
Neutrophil Elastase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
24 |
0.63 |
Binding ≤ 10μM
|
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
680 |
0.51 |
Binding ≤ 10μM
|
|
UROK-2-E |
Urokinase-type Plasminogen Activator (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
170 |
0.56 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.40 |
2.32 |
-9.96 |
0 |
6 |
0 |
80 |
251.629 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ELNE-3-E |
Neutrophil Elastase (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
45 |
0.69 |
Binding ≤ 10μM
|
|
KLKB1-1-E |
Plasma Kallikrein (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6200 |
0.49 |
Binding ≤ 10μM
|
|
THRB-5-E |
Prothrombin (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
4700 |
0.50 |
Binding ≤ 10μM
|
|
UROK-1-E |
Urokinase-type Plasminogen Activator (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1700 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.04 |
1.5 |
-6 |
0 |
3 |
0 |
34 |
265.11 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 4 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ELNE-3-E |
Neutrophil Elastase (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
104 |
0.65 |
Binding ≤ 10μM
|
|
KLKB1-1-E |
Plasma Kallikrein (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5700 |
0.49 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
2800 |
0.52 |
Binding ≤ 10μM
|
|
UROK-1-E |
Urokinase-type Plasminogen Activator (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
2600 |
0.52 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.14 |
1.67 |
-6.47 |
0 |
3 |
0 |
34 |
241.077 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 5 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ELNE-3-E |
Neutrophil Elastase (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
28 |
0.62 |
Binding ≤ 10μM
|
|
KLKB1-1-E |
Plasma Kallikrein (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6100 |
0.43 |
Binding ≤ 10μM
|
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
4300 |
0.44 |
Binding ≤ 10μM
|
|
UROK-1-E |
Urokinase-type Plasminogen Activator (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3100 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.39 |
2.9 |
-6.86 |
0 |
6 |
0 |
80 |
231.211 |
2 |
↓
|
|
|
|
Analogs
-
4891662
-
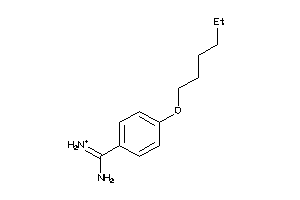
-
6535291
-
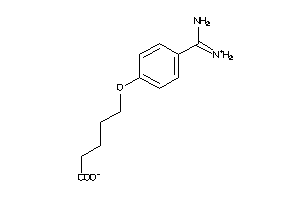
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.00 |
-4.42 |
-72.39 |
8 |
6 |
2 |
121 |
356.47 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.32 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
8 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.59 |
-9.99 |
-74.27 |
7 |
12 |
1 |
190 |
546.655 |
14 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.34 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
1000 |
0.23 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.05 |
3.88 |
-50.81 |
6 |
12 |
1 |
180 |
557.078 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 19 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
C1R-1-E |
Complement C1r (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1040 |
0.32 |
Binding ≤ 10μM |
|
C1S-1-E |
Complement C1s (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
140 |
0.37 |
Binding ≤ 10μM |
|
KLK1-1-E |
Kallikrein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
650 |
0.33 |
Binding ≤ 10μM |
|
PLMN-1-E |
Plasminogen (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2900 |
0.30 |
Binding ≤ 10μM
|
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
290 |
0.35 |
Binding ≤ 10μM |
|
TRY1-1-E |
Trypsin I (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
20 |
0.41 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.29 |
7.87 |
-80.13 |
9 |
7 |
2 |
142 |
349.394 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.99 |
5.81 |
-73.34 |
8 |
10 |
0 |
179 |
418.498 |
11 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.12 |
6.18 |
-100.16 |
9 |
10 |
1 |
183 |
419.506 |
10 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.12 |
4.82 |
-79.35 |
8 |
10 |
0 |
182 |
418.498 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.33 |
Binding ≤ 10μM
|
|
KLK1-1-E |
Kallikrein 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9700 |
0.19 |
Binding ≤ 10μM
|
|
THRB-4-E |
Prothrombin (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
5500 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.70 |
16.5 |
-37.08 |
2 |
5 |
1 |
51 |
514.024 |
6 |
↓
|
|
|
|
Analogs
-
14113515
-
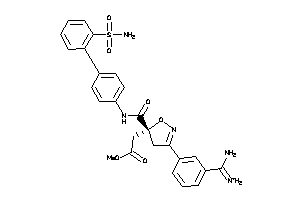
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3100 |
0.20 |
Binding ≤ 10μM
|
|
TRY1-1-E |
Trypsin I (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
98 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.42 |
5.3 |
-48.98 |
7 |
11 |
1 |
189 |
536.59 |
9 |
↓
|
|
|
|
Analogs
-
3920966
-
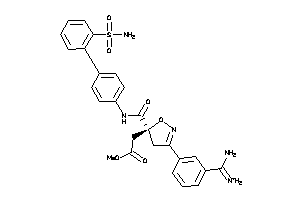
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.30 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3100 |
0.20 |
Binding ≤ 10μM
|
|
TRY1-1-E |
Trypsin I (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
98 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.42 |
5.28 |
-51.08 |
7 |
11 |
1 |
189 |
536.59 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.46 |
0.96 |
-5.24 |
0 |
4 |
0 |
44 |
266.3 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.19 |
-6.15 |
-84.43 |
8 |
6 |
2 |
121 |
472.181 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
5 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.43 |
-1.96 |
-16.95 |
2 |
8 |
0 |
101 |
452.824 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
91 |
0.66 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1425 |
0.55 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1425 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.08 |
-1.68 |
-13.7 |
7 |
7 |
0 |
134 |
229.265 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.21 |
-1.73 |
-13.45 |
7 |
7 |
0 |
137 |
229.265 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.08 |
-1.75 |
-14.11 |
7 |
7 |
0 |
134 |
229.265 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
203 |
0.62 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1215 |
0.55 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1215 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.65 |
1.07 |
-11.93 |
5 |
6 |
0 |
108 |
293.146 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.65 |
1.28 |
-41.01 |
4 |
6 |
-1 |
110 |
292.138 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.65 |
1.32 |
-38.67 |
4 |
6 |
-1 |
110 |
292.138 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
129 |
0.54 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1285 |
0.46 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1285 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.03 |
2.72 |
-13.47 |
5 |
6 |
0 |
108 |
264.31 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.03 |
2.92 |
-48.41 |
4 |
6 |
-1 |
110 |
263.302 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.03 |
2.96 |
-45.75 |
4 |
6 |
-1 |
110 |
263.302 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
2 |
0.39 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.01 |
6.4 |
-50.45 |
8 |
9 |
1 |
156 |
451.979 |
12 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.14 |
6.19 |
-54.4 |
8 |
9 |
1 |
159 |
451.979 |
11 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.14 |
6.75 |
-94.01 |
9 |
9 |
2 |
160 |
452.987 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.45 |
4.78 |
-43.04 |
5 |
8 |
1 |
127 |
453.544 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 44 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
95 |
0.70 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1350 |
0.59 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1350 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.97 |
-1.04 |
-14.13 |
6 |
6 |
0 |
125 |
214.25 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ELNE-1-E |
Neutrophil Elastase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
15 |
0.61 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
51 |
0.57 |
Binding ≤ 10μM
|
|
UROK-1-E |
Urokinase-type Plasminogen Activator (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
120 |
0.54 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.61 |
2.58 |
-9.44 |
0 |
5 |
0 |
61 |
264.668 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ELNE-1-E |
Neutrophil Elastase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
24 |
0.44 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
120 |
0.40 |
Binding ≤ 10μM
|
|
UROK-2-E |
Urokinase-type Plasminogen Activator (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1100 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.54 |
10.27 |
-6.67 |
0 |
4 |
0 |
52 |
322.339 |
3 |
↓
|
|
|
|
Analogs
-
6145401
-
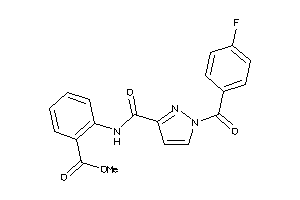
-
32373
-
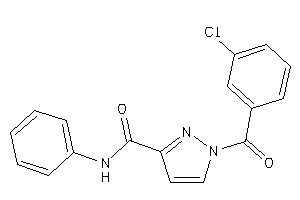
Draw
Identity
99%
90%
80%
70%
Vendors
And 1 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ELNE-3-E |
Neutrophil Elastase (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
39 |
0.47 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
390 |
0.41 |
Binding ≤ 10μM
|
|
UROK-1-E |
Urokinase-type Plasminogen Activator (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1500 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.99 |
0.73 |
-9.34 |
1 |
5 |
0 |
63 |
291.31 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 2 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ELNE-1-E |
Neutrophil Elastase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
107 |
0.54 |
Binding ≤ 10μM
|
|
KLKB1-1-E |
Plasma Kallikrein (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4400 |
0.42 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
2800 |
0.43 |
Binding ≤ 10μM
|
|
UROK-2-E |
Urokinase-type Plasminogen Activator (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1500 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.28 |
3.32 |
-8.08 |
0 |
6 |
0 |
80 |
249.201 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Vendors
And 9 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ELNE-1-E |
Neutrophil Elastase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
21 |
0.54 |
Binding ≤ 10μM
|
|
THRB-4-E |
Prothrombin (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
4800 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.71 |
5.24 |
-10.55 |
0 |
6 |
0 |
63 |
276.292 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
169 |
0.38 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1350 |
0.33 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1350 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.82 |
1.13 |
-13.32 |
2 |
8 |
0 |
110 |
361.427 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.82 |
0.54 |
-48.67 |
1 |
8 |
-1 |
107 |
360.419 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FA10-1-E |
Coagulation Factor X (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1 |
0.39 |
Binding ≤ 10μM
|
|
THRB-1-E |
Prothrombin (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3601 |
0.24 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.38 |
12.72 |
-54.29 |
3 |
5 |
1 |
63 |
473.062 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 9 More
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.97 |
-4.39 |
-75.96 |
5 |
10 |
0 |
154 |
471.521 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 8 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
220 |
0.67 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1230 |
0.59 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1230 |
0.59 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.76 |
1.71 |
-11.18 |
4 |
5 |
0 |
99 |
278.131 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
And 13 More
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
290 |
0.65 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1810 |
0.57 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1810 |
0.57 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.40 |
1.76 |
-13.17 |
4 |
5 |
0 |
99 |
213.262 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Vendors
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
THRB-8-E |
Prothrombin (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
170 |
0.56 |
Binding ≤ 10μM
|
|
TRY1-4-E |
Trypsin I (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1235 |
0.49 |
Binding ≤ 10μM
|
|
TRY2-3-E |
Trypsin II (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
1235 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.22 |
1.16 |
-17.26 |
5 |
9 |
0 |
154 |
259.247 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.22 |
1.37 |
-26.06 |
5 |
9 |
0 |
158 |
259.247 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.35 |
1.27 |
-48.76 |
6 |
9 |
1 |
158 |
260.255 |
4 |
↓
|
|