|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
44511690
-
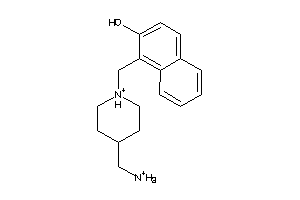
-
45663434
-
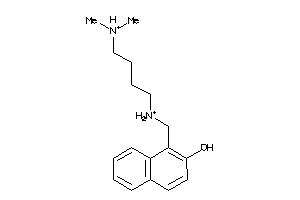
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
-0.43 |
-34.42 |
2 |
2 |
1 |
24 |
242.342 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.57 |
0.64 |
-47.76 |
0 |
5 |
-1 |
85 |
379.211 |
5 |
↓
|
|
|
|
Analogs
-
34646592
-
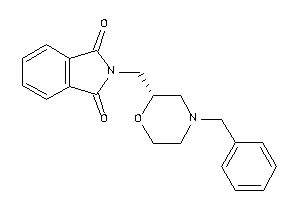
-
34646593
-
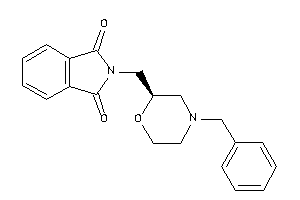
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.79 |
3.47 |
-45.71 |
2 |
5 |
1 |
65 |
247.274 |
2 |
↓
|
|
|
|
Analogs
-
34646592
-

-
34646593
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.79 |
3.41 |
-45.66 |
2 |
5 |
1 |
65 |
247.274 |
2 |
↓
|
|
|
|
Analogs
-
19855127
-
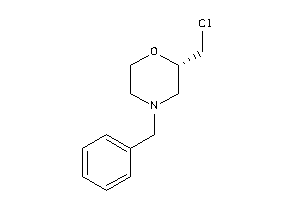
-
19855129
-
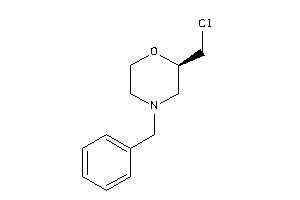
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.18 |
0.26 |
-40.47 |
1 |
2 |
1 |
14 |
226.727 |
3 |
↓
|
|
|
|
Analogs
-
44250011
-
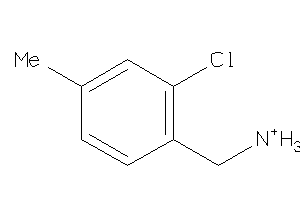
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.18 |
-1.67 |
-45.9 |
3 |
1 |
1 |
27 |
156.636 |
1 |
↓
|
|
|
|
Analogs
-
34599905
-
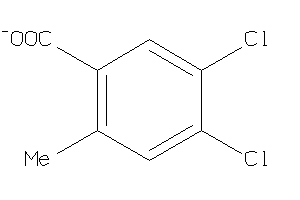
-
39386322
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.56 |
0.82 |
-44.73 |
0 |
2 |
-1 |
40 |
169.587 |
1 |
↓
|
|
|
|
Analogs
-
44225133
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.31 |
1.27 |
-51.76 |
0 |
2 |
-1 |
40 |
149.169 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.56 |
4.82 |
-41.09 |
0 |
5 |
-1 |
86 |
184.102 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.71 |
1.5 |
-50.72 |
0 |
2 |
-1 |
40 |
177.223 |
4 |
↓
|
|
|
|
Analogs
-
38416841
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.90 |
0.95 |
-47.91 |
0 |
2 |
-1 |
40 |
169.587 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.01 |
1.6 |
-43.42 |
0 |
0 |
-1 |
0 |
177.17 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.23 |
0.38 |
-50.58 |
2 |
4 |
-1 |
75 |
220.126 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.87 |
0.24 |
-37.94 |
0 |
1 |
-1 |
9 |
272.065 |
2 |
↓
|
|
|
|
Analogs
-
34334624
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.89 |
2.87 |
-49.85 |
2 |
3 |
-1 |
66 |
150.157 |
1 |
↓
|
|
|
|
Analogs
-
38236316
-
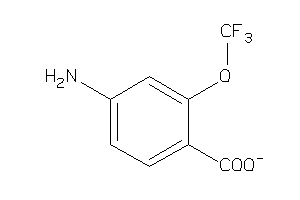
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.85 |
2.78 |
-51.54 |
0 |
3 |
-1 |
49 |
205.111 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.26 |
2.63 |
-41.28 |
2 |
2 |
1 |
29 |
123.179 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.26 |
2.91 |
-97.13 |
3 |
2 |
2 |
31 |
124.187 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.94 |
-0.04 |
-35.8 |
0 |
0 |
-1 |
0 |
161.608 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.78 |
-1.87 |
-43.87 |
3 |
1 |
1 |
27 |
191.081 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.22 |
-0.61 |
-43.58 |
0 |
1 |
-1 |
9 |
139.199 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
-2.09 |
-41.72 |
3 |
1 |
1 |
27 |
177.054 |
1 |
↓
|
|
|
|
Analogs
-
37094172
-
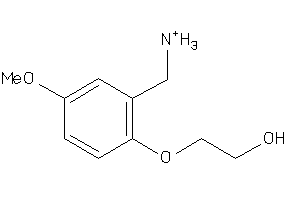
-
37094217
-
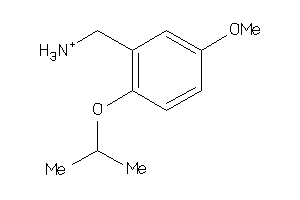
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.16 |
1.2 |
-40.09 |
3 |
3 |
1 |
46 |
168.216 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.89 |
3.09 |
-51.65 |
3 |
1 |
1 |
28 |
160.599 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
-2.14 |
-48.65 |
3 |
1 |
1 |
27 |
177.054 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.90 |
2.27 |
-102.81 |
7 |
10 |
1 |
161 |
500.58 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.92 |
3.71 |
-42.99 |
3 |
1 |
1 |
28 |
136.218 |
1 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.64 |
2.16 |
-39.86 |
3 |
1 |
1 |
28 |
193.089 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.80 |
0.43 |
-41.3 |
3 |
4 |
1 |
56 |
263.361 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.75 |
3.06 |
-30.71 |
4 |
2 |
1 |
52 |
135.19 |
1 |
↓
|
|
|
|
Analogs
-
44239226
-
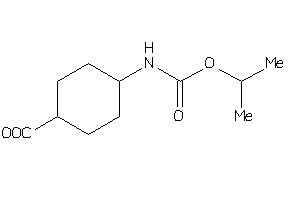
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
4.23 |
-51.73 |
1 |
5 |
-1 |
78 |
242.295 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
HCAR2-2-E |
HM74 Nicotinic Acid GPCR (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
900 |
0.94 |
Binding ≤ 10μM
|
|
OXDA-2-E |
D-amino-acid Oxidase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
910 |
0.94 |
Binding ≤ 10μM |
|
HCAR2-2-E |
HM74 Nicotinic Acid GPCR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
460 |
0.99 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.35 |
1.16 |
-43.14 |
1 |
4 |
-1 |
69 |
125.107 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.58 |
3.72 |
-40.57 |
0 |
3 |
-1 |
53 |
156.548 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.81 |
6.08 |
-46.25 |
0 |
4 |
-1 |
66 |
239.272 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.54 |
0.92 |
-8.15 |
1 |
4 |
0 |
63 |
240.28 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.51 |
1.07 |
-12.64 |
0 |
4 |
0 |
60 |
240.28 |
6 |
↓
|
|
|
|
Analogs
-
39134494
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.79 |
0.17 |
-30.14 |
4 |
3 |
1 |
65 |
142.207 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
-2.71 |
-46.83 |
3 |
1 |
1 |
27 |
198.698 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.56 |
2.43 |
-44.24 |
3 |
2 |
1 |
33 |
125.195 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.85 |
-1.29 |
-49.12 |
0 |
4 |
-1 |
62 |
260.294 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.37 |
1.38 |
-41.94 |
2 |
4 |
-1 |
79 |
137.118 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.37 |
1.86 |
-45.91 |
3 |
4 |
0 |
80 |
138.126 |
1 |
↓
|
|
|
|
Analogs
-
34525420
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.07 |
1.56 |
-35.8 |
1 |
3 |
1 |
24 |
281.379 |
4 |
↓
|
|
|
|
Analogs
-
34566573
-
-
34566575
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.03 |
-1.11 |
-42.11 |
3 |
2 |
1 |
44 |
150.201 |
2 |
↓
|
|
|
|
Analogs
-
34566573
-

-
34566575
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.03 |
-1.11 |
-41.64 |
3 |
2 |
1 |
44 |
150.201 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.34 |
0.46 |
-48.07 |
0 |
4 |
-1 |
66 |
129.12 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.40 |
4.01 |
-50.89 |
1 |
4 |
-1 |
73 |
188.162 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GABR1-2-E |
GABA-B Receptor 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
60 |
1.44 |
Binding ≤ 10μM |
|
GABR2-2-E |
GABA-B Receptor 2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
60 |
1.44 |
Binding ≤ 10μM |
|
GBRA1-7-E |
GABA Receptor Alpha-1 Subunit (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
10000 |
1.00 |
Binding ≤ 10μM
|
|
GBRA2-8-E |
GABA Receptor Alpha-2 Subunit (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
25 |
1.52 |
Binding ≤ 10μM
|
|
GBRA3-8-E |
GABA Receptor Alpha-3 Subunit (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
25 |
1.52 |
Binding ≤ 10μM
|
|
GBRA4-7-E |
GABA Receptor Alpha-4 Subunit (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
25 |
1.52 |
Binding ≤ 10μM
|
|
GBRA5-8-E |
GABA Receptor Alpha-5 Subunit (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
25 |
1.52 |
Binding ≤ 10μM
|
|
GBRA6-7-E |
GABA Receptor Alpha-6 Subunit (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
25 |
1.52 |
Binding ≤ 10μM
|
|
GBRB1-6-E |
GABA Receptor Beta-1 Subunit (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRB2-5-E |
GABA Receptor Beta-2 Subunit (cluster #5 Of 7), Eukaryotic |
Eukaryotes |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRB3-4-E |
GABA Receptor Beta-3 Subunit (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRD-3-E |
GABA Receptor Delta Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRE-3-E |
GABA Receptor Epsilon Subunit (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRG1-6-E |
GABA Receptor Gamma-1 Subunit (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRG2-6-E |
GABA Receptor Gamma-2 Subunit (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRG3-7-E |
GABA Receptor Gamma-3 Subunit (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRP-3-E |
GABA Receptor Pi Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRT-5-E |
GABA Receptor Theta Subunit (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
200 |
1.34 |
Binding ≤ 10μM
|
|
S6A11-1-E |
GABA Transporter 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7000 |
1.03 |
Binding ≤ 10μM
|
|
S6A13-1-E |
GABA Transporter 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
1.06 |
Binding ≤ 10μM
|
|
SC6A1-2-E |
GABA Transporter 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5000 |
1.06 |
Binding ≤ 10μM
|
|
GABR1-1-E |
GABA-B Receptor 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
1.15 |
Functional ≤ 10μM
|
|
GBRA1-5-E |
GABA Receptor Alpha-1 Subunit (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
1450 |
1.17 |
Functional ≤ 10μM
|
|
GBRA2-4-E |
GABA Receptor Alpha-2 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1450 |
1.17 |
Functional ≤ 10μM
|
|
GBRA3-4-E |
GABA Receptor Alpha-3 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1450 |
1.17 |
Functional ≤ 10μM
|
|
GBRA4-4-E |
GABA Receptor Alpha-4 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1450 |
1.17 |
Functional ≤ 10μM
|
|
GBRA5-4-E |
GABA Receptor Alpha-5 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
3000 |
1.10 |
Functional ≤ 10μM
|
|
GBRA6-4-E |
GABA Receptor Alpha-6 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
1500 |
1.16 |
Functional ≤ 10μM
|
|
GBRB3-2-E |
GABA Receptor Beta-3 Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3000 |
1.10 |
Functional ≤ 10μM
|
|
GBRG2-2-E |
GABA Receptor Gamma-2 Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3000 |
1.10 |
Functional ≤ 10μM
|
|
GBRR1-4-E |
GABA Receptor Rho-1 Subunit (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
800 |
1.22 |
Functional ≤ 10μM
|
|
GBRR2-2-E |
GABA Receptor Rho-2 Subunit (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1700 |
1.15 |
Functional ≤ 10μM
|
|
GBRR3-1-E |
GABA Receptor Rho-3 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1700 |
1.15 |
Functional ≤ 10μM
|
|
S6A11-2-E |
GABA Transporter 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3000 |
1.10 |
Functional ≤ 10μM
|
|
S6A12-1-E |
Betaine Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
1.10 |
Functional ≤ 10μM
|
|
S6A13-2-E |
GABA Transporter 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3000 |
1.10 |
Functional ≤ 10μM
|
|
SC6A1-1-E |
GABA Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
1.10 |
Functional ≤ 10μM
|
|
Z104301-3-O |
GABA-A Receptor; Anion Channel (cluster #3 Of 8), Other |
Other |
49 |
1.46 |
Binding ≤ 10μM
|
|
Z50597-3-O |
Rattus Norvegicus (cluster #3 Of 5), Other |
Other |
8600 |
1.01 |
Binding ≤ 10μM
|
|
Z50597-7-O |
Rattus Norvegicus (cluster #7 Of 12), Other |
Other |
70 |
1.43 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1_HUMAN |
P14867
|
GABA Receptor Alpha-1 Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRA2_HUMAN |
P47869
|
GABA Receptor Alpha-2 Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRA2_RAT |
P23576
|
GABA Receptor Alpha-2 Subunit, Rat |
22 |
1.53 |
Binding ≤ 1μM
|
|
GBRA3_HUMAN |
P34903
|
GABA Receptor Alpha-3 Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRA4_HUMAN |
P48169
|
GABA Receptor Alpha-4 Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRA5_HUMAN |
P31644
|
GABA Receptor Alpha-5 Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRA6_HUMAN |
Q16445
|
GABA Receptor Alpha-6 Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRB1_HUMAN |
P18505
|
GABA Receptor Beta-1 Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRB2_HUMAN |
P47870
|
GABA Receptor Beta-2 Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRB3_HUMAN |
P28472
|
GABA Receptor Beta-3 Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRD_HUMAN |
O14764
|
GABA Receptor Delta Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRE_HUMAN |
P78334
|
GABA Receptor Epsilon Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRG1_RAT |
P23574
|
GABA Receptor Gamma-1 Subunit, Rat |
33 |
1.50 |
Binding ≤ 1μM
|
|
GBRG1_HUMAN |
Q8N1C3
|
GABA Receptor Gamma-1 Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRG2_HUMAN |
P18507
|
GABA Receptor Gamma-2 Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRG3_HUMAN |
Q99928
|
GABA Receptor Gamma-3 Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRP_HUMAN |
O00591
|
GABA Receptor Pi Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
GBRT_HUMAN |
Q9UN88
|
GABA Receptor Theta Subunit, Human |
200 |
1.34 |
Binding ≤ 1μM
|
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
100 |
1.40 |
Binding ≤ 1μM
|
|
GABR1_RAT |
Q9Z0U4
|
GABA-B Receptor 1, Rat |
17 |
1.55 |
Binding ≤ 1μM
|
|
GABR1_HUMAN |
Q9UBS5
|
GABA-B Receptor 1, Human |
25 |
1.52 |
Binding ≤ 1μM
|
|
GABR2_RAT |
O88871
|
GABA-B Receptor 2, Rat |
17 |
1.55 |
Binding ≤ 1μM
|
|
GABR2_HUMAN |
O75899
|
GABA-B Receptor 2, Human |
25 |
1.52 |
Binding ≤ 1μM
|
|
GBRA1_HUMAN |
P14867
|
GABA Receptor Alpha-1 Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRA1_RAT |
P62813
|
GABA Receptor Alpha-1 Subunit, Rat |
10000 |
1.00 |
Binding ≤ 10μM
|
|
GBRA2_HUMAN |
P47869
|
GABA Receptor Alpha-2 Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRA2_RAT |
P23576
|
GABA Receptor Alpha-2 Subunit, Rat |
22 |
1.53 |
Binding ≤ 10μM
|
|
GBRA3_HUMAN |
P34903
|
GABA Receptor Alpha-3 Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRA4_HUMAN |
P48169
|
GABA Receptor Alpha-4 Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRA5_HUMAN |
P31644
|
GABA Receptor Alpha-5 Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRA6_HUMAN |
Q16445
|
GABA Receptor Alpha-6 Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRB1_HUMAN |
P18505
|
GABA Receptor Beta-1 Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRB2_HUMAN |
P47870
|
GABA Receptor Beta-2 Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRB3_HUMAN |
P28472
|
GABA Receptor Beta-3 Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRD_HUMAN |
O14764
|
GABA Receptor Delta Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRE_HUMAN |
P78334
|
GABA Receptor Epsilon Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRG1_HUMAN |
Q8N1C3
|
GABA Receptor Gamma-1 Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRG1_RAT |
P23574
|
GABA Receptor Gamma-1 Subunit, Rat |
33 |
1.50 |
Binding ≤ 10μM
|
|
GBRG2_HUMAN |
P18507
|
GABA Receptor Gamma-2 Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRG3_HUMAN |
Q99928
|
GABA Receptor Gamma-3 Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRP_HUMAN |
O00591
|
GABA Receptor Pi Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
GBRT_HUMAN |
Q9UN88
|
GABA Receptor Theta Subunit, Human |
200 |
1.34 |
Binding ≤ 10μM
|
|
SC6A1_HUMAN |
P30531
|
GABA Transporter 1, Human |
5000 |
1.06 |
Binding ≤ 10μM
|
|
SC6A1_RAT |
P23978
|
GABA Transporter 1, Rat |
1620 |
1.16 |
Binding ≤ 10μM
|
|
S6A13_RAT |
P31646
|
GABA Transporter 2, Rat |
5000 |
1.06 |
Binding ≤ 10μM
|
|
S6A11_HUMAN |
P48066
|
GABA Transporter 3, Human |
7000 |
1.03 |
Binding ≤ 10μM
|
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
100 |
1.40 |
Binding ≤ 10μM
|
|
GABR1_RAT |
Q9Z0U4
|
GABA-B Receptor 1, Rat |
17 |
1.55 |
Binding ≤ 10μM
|
|
GABR1_HUMAN |
Q9UBS5
|
GABA-B Receptor 1, Human |
25 |
1.52 |
Binding ≤ 10μM
|
|
GABR2_RAT |
O88871
|
GABA-B Receptor 2, Rat |
17 |
1.55 |
Binding ≤ 10μM
|
|
GABR2_HUMAN |
O75899
|
GABA-B Receptor 2, Human |
25 |
1.52 |
Binding ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
8600 |
1.01 |
Binding ≤ 10μM
|
|
S6A12_RAT |
P48056
|
Betaine Transporter, Rat |
2000 |
1.14 |
Functional ≤ 10μM
|
|
GBRA1_HUMAN |
P14867
|
GABA Receptor Alpha-1 Subunit, Human |
1450 |
1.17 |
Functional ≤ 10μM
|
|
GBRA2_HUMAN |
P47869
|
GABA Receptor Alpha-2 Subunit, Human |
1450 |
1.17 |
Functional ≤ 10μM
|
|
GBRA3_HUMAN |
P34903
|
GABA Receptor Alpha-3 Subunit, Human |
1450 |
1.17 |
Functional ≤ 10μM
|
|
GBRA4_HUMAN |
P48169
|
GABA Receptor Alpha-4 Subunit, Human |
1450 |
1.17 |
Functional ≤ 10μM
|
|
GBRA5_HUMAN |
P31644
|
GABA Receptor Alpha-5 Subunit, Human |
1450 |
1.17 |
Functional ≤ 10μM
|
|
GBRA6_HUMAN |
Q16445
|
GABA Receptor Alpha-6 Subunit, Human |
1450 |
1.17 |
Functional ≤ 10μM
|
|
GBRB3_HUMAN |
P28472
|
GABA Receptor Beta-3 Subunit, Human |
1500 |
1.16 |
Functional ≤ 10μM
|
|
GBRG2_HUMAN |
P18507
|
GABA Receptor Gamma-2 Subunit, Human |
1500 |
1.16 |
Functional ≤ 10μM
|
|
GBRR1_HUMAN |
P24046
|
GABA Receptor Rho-1 Subunit, Human |
1190 |
1.18 |
Functional ≤ 10μM
|
|
GBRR2_HUMAN |
P28476
|
GABA Receptor Rho-2 Subunit, Human |
1700 |
1.15 |
Functional ≤ 10μM
|
|
GBRR3_HUMAN |
A8MPY1
|
GABA Receptor Rho-3 Subunit, Human |
1700 |
1.15 |
Functional ≤ 10μM
|
|
SC6A1_RAT |
P23978
|
GABA Transporter 1, Rat |
2000 |
1.14 |
Functional ≤ 10μM
|
|
S6A13_RAT |
P31646
|
GABA Transporter 2, Rat |
2000 |
1.14 |
Functional ≤ 10μM
|
|
S6A11_RAT |
P31647
|
GABA Transporter 3, Rat |
2000 |
1.14 |
Functional ≤ 10μM
|
|
GABR1_HUMAN |
Q9UBS5
|
GABA-B Receptor 1, Human |
1700 |
1.15 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
2000 |
1.14 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.10 |
0.98 |
-62.79 |
3 |
3 |
0 |
68 |
103.121 |
3 |
↓
|
|
|
|
Analogs
-
37005495
-
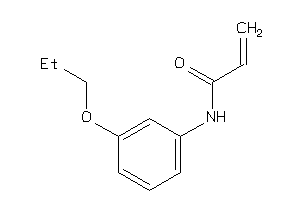
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.26 |
1.79 |
-52.2 |
4 |
4 |
1 |
66 |
209.269 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.65 |
3.85 |
-45.53 |
0 |
3 |
-1 |
53 |
136.13 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.65 |
4.3 |
-45.37 |
1 |
3 |
0 |
54 |
137.138 |
1 |
↓
|
|