|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.66 |
3.68 |
-10.9 |
0 |
3 |
0 |
43 |
428.657 |
10 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
-0.32 |
-5.28 |
1 |
2 |
0 |
37 |
290.447 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
-0.29 |
-6.05 |
1 |
2 |
0 |
37 |
290.447 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
-0.28 |
-5.32 |
1 |
2 |
0 |
37 |
290.447 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
-0.36 |
-5.94 |
1 |
2 |
0 |
37 |
290.447 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.61 |
9.24 |
-7.63 |
0 |
3 |
0 |
33 |
324.811 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.61 |
10.11 |
-30.26 |
1 |
3 |
1 |
34 |
325.819 |
3 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
4214283
-
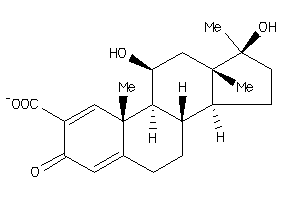
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.51 |
-0.94 |
-19.39 |
2 |
4 |
0 |
74 |
344.451 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.85 |
-0.5 |
-9.84 |
0 |
3 |
0 |
32 |
302.736 |
1 |
↓
|
|
|
|
Analogs
-
103270
-
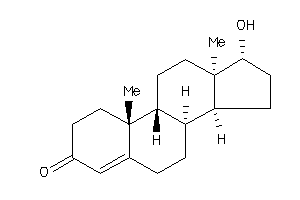
-
490793
-
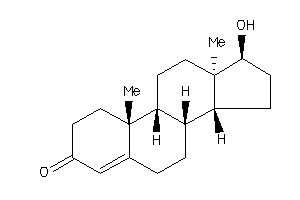
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.17 |
0.35 |
-6.53 |
1 |
2 |
0 |
37 |
316.485 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.87 |
-1.25 |
-3.08 |
2 |
2 |
0 |
40 |
165.403 |
1 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.27 |
-1.73 |
-8.63 |
0 |
3 |
0 |
32 |
318.829 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
-3.46 |
-8.43 |
1 |
4 |
0 |
54 |
316.158 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.07 |
3.48 |
-8.87 |
1 |
4 |
0 |
42 |
349.217 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.07 |
3.48 |
-8.88 |
1 |
4 |
0 |
42 |
349.217 |
1 |
↓
|
|
|
|
Analogs
-
2011625
-
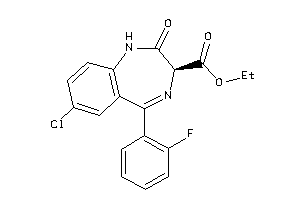
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.83 |
-1 |
-9.85 |
1 |
5 |
0 |
67 |
360.772 |
4 |
↓
|
|
|
|
Analogs
-
601273
-
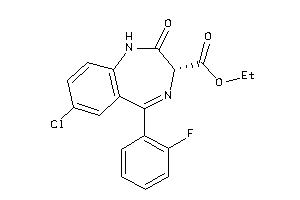
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.83 |
-1 |
-9.86 |
1 |
5 |
0 |
67 |
360.772 |
4 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
31560325
-
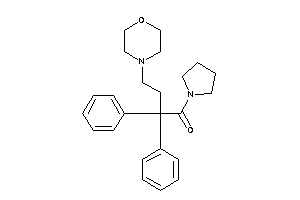
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.95 |
10.39 |
-7.01 |
0 |
4 |
0 |
33 |
392.543 |
6 |
↓
|
|
|
|
Analogs
-
31560325
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.95 |
8.91 |
-9.07 |
0 |
4 |
0 |
33 |
392.543 |
6 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
4.81 |
-3.19 |
2 |
2 |
0 |
40 |
290.447 |
0 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.81 |
6.4 |
-8.56 |
1 |
4 |
0 |
59 |
330.472 |
0 |
↓
|
|
|
|
Analogs
-
34327160
-
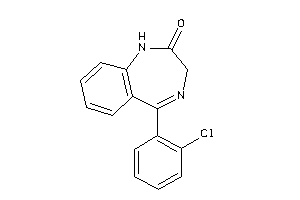
-
39296227
-
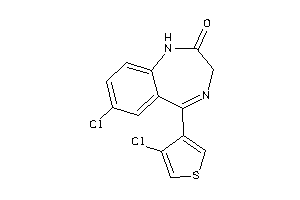
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRA5-6-E |
GABA Receptor Alpha-5 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRA6-2-E |
GABA Receptor Alpha-6 Subunit (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRB2-1-E |
GABA Receptor Beta-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRB3-1-E |
GABA Receptor Beta-3 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRD-1-E |
GABA Receptor Delta Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRE-1-E |
GABA Receptor Epsilon Subunit (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRG1-1-E |
GABA Receptor Gamma-1 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRG2-1-E |
GABA Receptor Gamma-2 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRG3-2-E |
GABA Receptor Gamma-3 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRP-1-E |
GABA Receptor Pi Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
|
GBRT-2-E |
GABA Receptor Theta Subunit (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
3 |
0.60 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1_HUMAN |
P14867
|
GABA Receptor Alpha-1 Subunit, Human |
2.8 |
0.60 |
Binding ≤ 1μM
|
|
GBRA2_HUMAN |
P47869
|
GABA Receptor Alpha-2 Subunit, Human |
2.8 |
0.60 |
Binding ≤ 1μM
|
|
GBRA3_HUMAN |
P34903
|
GABA Receptor Alpha-3 Subunit, Human |
2.8 |
0.60 |
Binding ≤ 1μM
|
|
GBRA4_HUMAN |
P48169
|
GABA Receptor Alpha-4 Subunit, Human |
2.8 |
0.60 |
Binding ≤ 1μM
|
|
GBRA5_HUMAN |
P31644
|
GABA Receptor Alpha-5 Subunit, Human |
2.8 |
0.60 |
Binding ≤ 1μM
|
|
GBRA6_HUMAN |
Q16445
|
GABA Receptor Alpha-6 Subunit, Human |
2.8 |
0.60 |
Binding ≤ 1μM
|
|
GBRB1_HUMAN |
P18505
|
GABA Receptor Beta-1 Subunit, Human |
2.8 |
0.60 |
Binding ≤ 1μM
|
|
GBRB2_HUMAN |
P47870
|
GABA Receptor Beta-2 Subunit, Human |
2.8 |
0.60 |
Binding ≤ 1μM
|
|
GBRB3_HUMAN |
P28472
|
GABA Receptor Beta-3 Subunit, Human |
2.8 |
0.60 |
Binding ≤ 1μM
|
|
GBRD_HUMAN |
O14764
|
GABA Receptor Delta Subunit, Human |
2.8 |
0.60 |
Binding ≤ 1μM
|
|
GBRE_HUMAN |
P78334
|
GABA Receptor Epsilon Subunit, Human |
2.8 |
0.60 |
Binding ≤ 1μM
|
|
GBRG1_HUMAN |
Q8N1C3
|
GABA Receptor Gamma-1 Subunit, Human |
2.8 |
0.60 |
Binding ≤ 1μM
|
|
GBRG2_HUMAN |
P18507
|
GABA Receptor Gamma-2 Subunit, Human |
2.8 |
0.60 |
Binding ≤ 1μM
|
|
GBRG3_HUMAN |
Q99928
|
GABA Receptor Gamma-3 Subunit, Human |
2.8 |
0.60 |
Binding ≤ 1μM
|
|
GBRP_HUMAN |
O00591
|
GABA Receptor Pi Subunit, Human |
2.8 |
0.60 |
Binding ≤ 1μM
|
|
GBRT_HUMAN |
Q9UN88
|
GABA Receptor Theta Subunit, Human |
2.8 |
0.60 |
Binding ≤ 1μM
|
|
GBRA1_HUMAN |
P14867
|
GABA Receptor Alpha-1 Subunit, Human |
2.8 |
0.60 |
Binding ≤ 10μM
|
|
GBRA2_HUMAN |
P47869
|
GABA Receptor Alpha-2 Subunit, Human |
2.8 |
0.60 |
Binding ≤ 10μM
|
|
GBRA3_HUMAN |
P34903
|
GABA Receptor Alpha-3 Subunit, Human |
2.8 |
0.60 |
Binding ≤ 10μM
|
|
GBRA4_HUMAN |
P48169
|
GABA Receptor Alpha-4 Subunit, Human |
2.8 |
0.60 |
Binding ≤ 10μM
|
|
GBRA5_HUMAN |
P31644
|
GABA Receptor Alpha-5 Subunit, Human |
2.8 |
0.60 |
Binding ≤ 10μM
|
|
GBRA6_HUMAN |
Q16445
|
GABA Receptor Alpha-6 Subunit, Human |
2.8 |
0.60 |
Binding ≤ 10μM
|
|
GBRB1_HUMAN |
P18505
|
GABA Receptor Beta-1 Subunit, Human |
2.8 |
0.60 |
Binding ≤ 10μM
|
|
GBRB2_HUMAN |
P47870
|
GABA Receptor Beta-2 Subunit, Human |
2.8 |
0.60 |
Binding ≤ 10μM
|
|
GBRB3_HUMAN |
P28472
|
GABA Receptor Beta-3 Subunit, Human |
2.8 |
0.60 |
Binding ≤ 10μM
|
|
GBRD_HUMAN |
O14764
|
GABA Receptor Delta Subunit, Human |
2.8 |
0.60 |
Binding ≤ 10μM
|
|
GBRE_HUMAN |
P78334
|
GABA Receptor Epsilon Subunit, Human |
2.8 |
0.60 |
Binding ≤ 10μM
|
|
GBRG1_HUMAN |
Q8N1C3
|
GABA Receptor Gamma-1 Subunit, Human |
2.8 |
0.60 |
Binding ≤ 10μM
|
|
GBRG2_HUMAN |
P18507
|
GABA Receptor Gamma-2 Subunit, Human |
2.8 |
0.60 |
Binding ≤ 10μM
|
|
GBRG3_HUMAN |
Q99928
|
GABA Receptor Gamma-3 Subunit, Human |
2.8 |
0.60 |
Binding ≤ 10μM
|
|
GBRP_HUMAN |
O00591
|
GABA Receptor Pi Subunit, Human |
2.8 |
0.60 |
Binding ≤ 10μM
|
|
GBRT_HUMAN |
Q9UN88
|
GABA Receptor Theta Subunit, Human |
2.8 |
0.60 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.49 |
5.58 |
-8.46 |
1 |
3 |
0 |
41 |
305.164 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.49 |
6.5 |
-34.12 |
2 |
3 |
1 |
43 |
306.172 |
1 |
↓
|
|
|
|
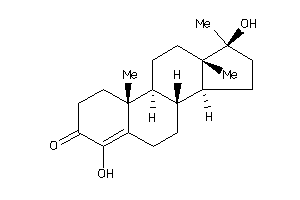
Analogs
-
4217232
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
4.87 |
-8.36 |
2 |
3 |
0 |
58 |
290.403 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.40 |
5.88 |
-16.84 |
0 |
3 |
0 |
27 |
188.23 |
1 |
↓
|
|
|
|
|
|
|
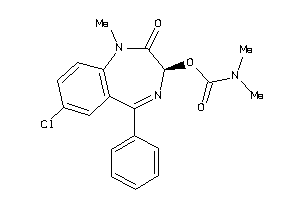
Analogs
-
2008504
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.59 |
-1.65 |
-15.17 |
0 |
6 |
0 |
62 |
371.824 |
3 |
↓
|
|
|
|
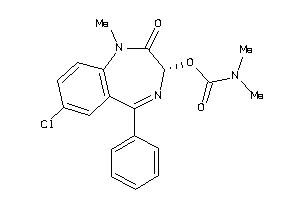
Analogs
-
601288
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.59 |
-1.84 |
-15.77 |
0 |
6 |
0 |
62 |
371.824 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.78 |
-1.67 |
-12.54 |
2 |
4 |
0 |
58 |
293.407 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.74 |
-0.67 |
-5.46 |
2 |
5 |
0 |
75 |
184.195 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.04 |
-3.18 |
-35.95 |
1 |
5 |
-1 |
82 |
183.187 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.68 |
3.69 |
-9.33 |
1 |
4 |
0 |
42 |
377.213 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.68 |
3.69 |
-9.35 |
1 |
4 |
0 |
42 |
377.213 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.96 |
9.25 |
-7.5 |
0 |
4 |
0 |
39 |
353.462 |
8 |
↓
|
|
|
|
Analogs
-
103270
-

-
490793
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.17 |
0.45 |
-6.88 |
1 |
2 |
0 |
37 |
316.485 |
0 |
↓
|
|
|
|
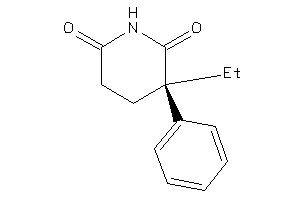
Analogs
-
1530797
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.69 |
4.27 |
-9.63 |
1 |
3 |
0 |
46 |
217.268 |
2 |
↓
|
|
|
|
Analogs
-
1530796
-
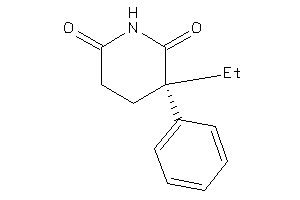
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.69 |
-0.06 |
-9.52 |
1 |
3 |
0 |
46 |
217.268 |
2 |
↓
|
|