|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
270 |
0.34 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
26 |
0.39 |
Binding ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
35 |
0.39 |
Binding ≤ 10μM
|
|
AA3R-1-E |
Adenosine Receptor A3 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
660 |
0.32 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R_HUMAN |
P30542
|
Adenosine A1 Receptor, Human |
266 |
0.34 |
Binding ≤ 1μM
|
|
AA2AR_RAT |
P30543
|
Adenosine A2a Receptor, Rat |
26 |
0.39 |
Binding ≤ 1μM
|
|
AA2AR_HUMAN |
P29274
|
Adenosine A2a Receptor, Human |
2.7 |
0.44 |
Binding ≤ 1μM
|
|
AA2BR_HUMAN |
P29275
|
Adenosine A2b Receptor, Human |
35 |
0.39 |
Binding ≤ 1μM
|
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
660 |
0.32 |
Binding ≤ 1μM
|
|
AA1R_HUMAN |
P30542
|
Adenosine A1 Receptor, Human |
266 |
0.34 |
Binding ≤ 10μM
|
|
AA2AR_RAT |
P30543
|
Adenosine A2a Receptor, Rat |
26 |
0.39 |
Binding ≤ 10μM
|
|
AA2AR_HUMAN |
P29274
|
Adenosine A2a Receptor, Human |
2.7 |
0.44 |
Binding ≤ 10μM
|
|
AA2BR_HUMAN |
P29275
|
Adenosine A2b Receptor, Human |
35 |
0.39 |
Binding ≤ 10μM
|
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
660 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.35 |
7.23 |
-47.09 |
2 |
9 |
1 |
94 |
368.421 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.35 |
4.84 |
-18.78 |
1 |
9 |
0 |
92 |
367.413 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1730 |
0.28 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
14 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.79 |
8.67 |
-46.92 |
2 |
9 |
1 |
94 |
396.475 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.79 |
6.28 |
-18.72 |
1 |
9 |
0 |
92 |
395.467 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
850 |
0.30 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
135 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.57 |
8.05 |
-47.09 |
2 |
9 |
1 |
94 |
382.448 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.57 |
5.66 |
-18.88 |
1 |
9 |
0 |
92 |
381.44 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
390 |
0.31 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
14 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.79 |
8.82 |
-47.01 |
2 |
9 |
1 |
94 |
396.475 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.79 |
6.41 |
-18.9 |
1 |
9 |
0 |
92 |
395.467 |
5 |
↓
|
|
|
|
Analogs
-
23360551
-
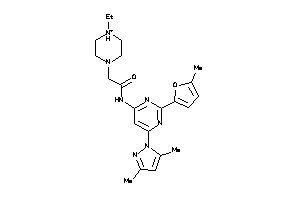
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
850 |
0.28 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
131 |
0.32 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
950 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.01 |
9.41 |
-47.05 |
2 |
9 |
1 |
94 |
410.502 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.01 |
6.99 |
-18.97 |
1 |
9 |
0 |
92 |
409.494 |
5 |
↓
|
|
|
|
Analogs
-
23360520
-
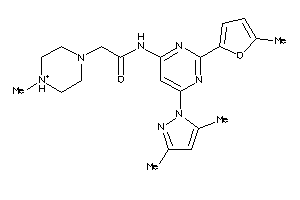
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1880 |
0.26 |
Binding ≤ 10μM
|
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
270 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.39 |
10.08 |
-45.78 |
2 |
9 |
1 |
94 |
424.529 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.39 |
7.8 |
-18.86 |
1 |
9 |
0 |
92 |
423.521 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.21 |
8.98 |
-37.95 |
1 |
9 |
0 |
100 |
423.521 |
6 |
↓
|
|