|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.43 |
-0.24 |
-9.13 |
1 |
5 |
0 |
55 |
309.417 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.43 |
-0.12 |
-40.59 |
2 |
5 |
1 |
56 |
310.425 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.22 |
11.27 |
-34.15 |
2 |
5 |
1 |
61 |
310.425 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.22 |
11.8 |
-89.92 |
3 |
5 |
2 |
62 |
311.433 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.86 |
11.58 |
-31.68 |
2 |
6 |
1 |
74 |
345.43 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.42 |
0.19 |
-76.96 |
3 |
6 |
2 |
75 |
346.438 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.42 |
0.49 |
-156.29 |
4 |
6 |
3 |
76 |
347.446 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.72 |
-0.92 |
-11.08 |
1 |
6 |
0 |
68 |
330.395 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.72 |
-0.81 |
-43.92 |
2 |
6 |
1 |
69 |
331.403 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.72 |
-0.81 |
-41.75 |
2 |
6 |
1 |
69 |
331.403 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.91 |
11.76 |
-34.66 |
2 |
5 |
1 |
61 |
310.425 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.91 |
12.21 |
-89.93 |
3 |
5 |
2 |
62 |
311.433 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.34 |
-0.02 |
-43.32 |
2 |
6 |
1 |
60 |
325.44 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.34 |
0.09 |
-81.99 |
3 |
6 |
2 |
61 |
326.448 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.79 |
11.66 |
-29.32 |
2 |
5 |
1 |
61 |
310.425 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.50 |
1.05 |
-75.8 |
3 |
5 |
2 |
62 |
311.433 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
-0.52 |
-13.34 |
1 |
5 |
0 |
55 |
309.417 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.06 |
-0.21 |
-32.6 |
2 |
5 |
1 |
56 |
310.425 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.97 |
6.85 |
-36.91 |
3 |
6 |
1 |
81 |
298.37 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.97 |
7.31 |
-92.57 |
4 |
6 |
2 |
82 |
299.378 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.92 |
-0.07 |
-46.96 |
2 |
6 |
1 |
60 |
339.467 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.92 |
0.24 |
-76.29 |
3 |
6 |
2 |
61 |
340.475 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.58 |
7.79 |
-30.91 |
3 |
6 |
1 |
81 |
312.397 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.58 |
8.15 |
-95.53 |
4 |
6 |
2 |
82 |
313.405 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
FAK1-1-E |
Focal Adhesion Kinase 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
38 |
0.29 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.25 |
12.6 |
-56.17 |
3 |
10 |
1 |
104 |
491.572 |
10 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.25 |
10.13 |
-19.12 |
2 |
10 |
0 |
103 |
490.564 |
10 |
↓
|
|
|
|
Analogs
-
38429336
-
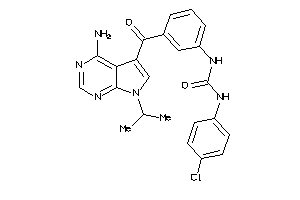
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.82 |
10.7 |
-14.16 |
4 |
9 |
0 |
124 |
513.385 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.03 |
4 |
-56.28 |
1 |
5 |
-1 |
82 |
196.573 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.45 |
8.39 |
-11.26 |
1 |
6 |
0 |
63 |
324.384 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.45 |
8.86 |
-36.71 |
2 |
6 |
1 |
65 |
325.392 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.45 |
8.39 |
-11.4 |
1 |
6 |
0 |
63 |
324.384 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.45 |
8.86 |
-35.63 |
2 |
6 |
1 |
65 |
325.392 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.96 |
11.92 |
-56.05 |
1 |
6 |
-1 |
85 |
369.832 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.96 |
9.94 |
-10.68 |
2 |
6 |
0 |
82 |
370.84 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.96 |
12.23 |
-55.07 |
1 |
6 |
-1 |
85 |
369.832 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.96 |
10.43 |
-14.11 |
2 |
6 |
0 |
82 |
370.84 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.51 |
10.99 |
-11.32 |
1 |
6 |
0 |
71 |
364.449 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.51 |
11.46 |
-36.38 |
2 |
6 |
1 |
72 |
365.457 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.51 |
11 |
-11.45 |
1 |
6 |
0 |
71 |
364.449 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.51 |
11.47 |
-36.18 |
2 |
6 |
1 |
72 |
365.457 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.57 |
11.24 |
-12.66 |
1 |
5 |
0 |
62 |
374.366 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.57 |
11.71 |
-38.41 |
2 |
5 |
1 |
63 |
375.374 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.57 |
11.25 |
-12.49 |
1 |
5 |
0 |
62 |
374.366 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.57 |
11.72 |
-38.52 |
2 |
5 |
1 |
63 |
375.374 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
11.2 |
-13.31 |
1 |
6 |
0 |
71 |
364.449 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.47 |
11.61 |
-37.81 |
2 |
6 |
1 |
72 |
365.457 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.47 |
11.2 |
-13.02 |
1 |
6 |
0 |
71 |
364.449 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.47 |
11.61 |
-37.82 |
2 |
6 |
1 |
72 |
365.457 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.98 |
5.1 |
-65.28 |
5 |
7 |
1 |
102 |
317.417 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.98 |
4.85 |
-13.52 |
4 |
7 |
0 |
100 |
316.409 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.98 |
5.57 |
-104.07 |
6 |
7 |
2 |
103 |
318.425 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.98 |
5.05 |
-57.61 |
5 |
7 |
1 |
102 |
317.417 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.98 |
4.79 |
-14.85 |
4 |
7 |
0 |
100 |
316.409 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.98 |
5.52 |
-100 |
6 |
7 |
2 |
103 |
318.425 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AKT1-2-E |
RAC-alpha Serine/threonine-protein Kinase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
765 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.10 |
3.63 |
-64.62 |
5 |
7 |
1 |
102 |
289.363 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.10 |
3.37 |
-14.25 |
4 |
7 |
0 |
100 |
288.355 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.10 |
4.09 |
-102.94 |
6 |
7 |
2 |
103 |
290.371 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AKT1-2-E |
RAC-alpha Serine/threonine-protein Kinase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
765 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.10 |
3.57 |
-57.15 |
5 |
7 |
1 |
102 |
289.363 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.10 |
3.31 |
-15.53 |
4 |
7 |
0 |
100 |
288.355 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.10 |
4.04 |
-99.06 |
6 |
7 |
2 |
103 |
290.371 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AKT1-2-E |
RAC-alpha Serine/threonine-protein Kinase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
4000 |
0.38 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AKT1_HUMAN |
P31749
|
Serine/threonine-protein Kinase AKT, Human |
4000 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.43 |
3.21 |
-17.32 |
2 |
7 |
0 |
91 |
295.368 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.43 |
3.68 |
-41.36 |
3 |
7 |
1 |
92 |
296.376 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.43 |
3.22 |
-17.42 |
2 |
7 |
0 |
91 |
295.368 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.43 |
3.69 |
-40.91 |
3 |
7 |
1 |
92 |
296.376 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AKT1-2-E |
RAC-alpha Serine/threonine-protein Kinase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
600 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.97 |
6.41 |
-15.43 |
2 |
7 |
0 |
91 |
357.439 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.97 |
6.88 |
-39.42 |
3 |
7 |
1 |
92 |
358.447 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.97 |
6.46 |
-15.03 |
2 |
7 |
0 |
91 |
357.439 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.97 |
6.93 |
-39.18 |
3 |
7 |
1 |
92 |
358.447 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AKT1-2-E |
RAC-alpha Serine/threonine-protein Kinase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
2400 |
0.34 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AKT1_HUMAN |
P31749
|
Serine/threonine-protein Kinase AKT, Human |
2400 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.46 |
10.08 |
-50.32 |
3 |
5 |
1 |
61 |
308.409 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.46 |
8.89 |
-8.65 |
2 |
5 |
0 |
57 |
307.401 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.46 |
10.1 |
-50.25 |
3 |
5 |
1 |
61 |
308.409 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.46 |
8.93 |
-9.66 |
2 |
5 |
0 |
57 |
307.401 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AKT1-2-E |
RAC-alpha Serine/threonine-protein Kinase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
38 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.71 |
6.81 |
-73.22 |
5 |
7 |
1 |
102 |
369.424 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.71 |
6.55 |
-16.52 |
4 |
7 |
0 |
100 |
368.416 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.71 |
7.28 |
-113.18 |
6 |
7 |
2 |
103 |
370.432 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AKT1-2-E |
RAC-alpha Serine/threonine-protein Kinase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
38 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.71 |
6.74 |
-65.46 |
5 |
7 |
1 |
102 |
369.424 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.71 |
6.49 |
-18.12 |
4 |
7 |
0 |
100 |
368.416 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.71 |
7.21 |
-109.24 |
6 |
7 |
2 |
103 |
370.432 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AKT1-2-E |
RAC-alpha Serine/threonine-protein Kinase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.44 |
Binding ≤ 10μM
|
|
AKT1-2-E |
RAC-alpha Serine/threonine-protein Kinase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.74 |
6.81 |
-70.3 |
5 |
7 |
1 |
102 |
369.424 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.74 |
6.55 |
-14.36 |
4 |
7 |
0 |
100 |
368.416 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.74 |
7.28 |
-110.64 |
6 |
7 |
2 |
103 |
370.432 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AKT1-2-E |
RAC-alpha Serine/threonine-protein Kinase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.74 |
6.75 |
-63 |
5 |
7 |
1 |
102 |
369.424 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.74 |
6.49 |
-15.73 |
4 |
7 |
0 |
100 |
368.416 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.74 |
7.21 |
-106.78 |
6 |
7 |
2 |
103 |
370.432 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AKT1-2-E |
RAC-alpha Serine/threonine-protein Kinase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
61 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.79 |
4.01 |
-64.45 |
5 |
7 |
1 |
102 |
343.333 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.79 |
3.77 |
-11.56 |
4 |
7 |
0 |
100 |
342.325 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.79 |
4.48 |
-104.7 |
6 |
7 |
2 |
103 |
344.341 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AKT1-2-E |
RAC-alpha Serine/threonine-protein Kinase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
61 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.79 |
3.97 |
-58.26 |
5 |
7 |
1 |
102 |
343.333 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.79 |
3.7 |
-12.34 |
4 |
7 |
0 |
100 |
342.325 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.79 |
4.44 |
-101.39 |
6 |
7 |
2 |
103 |
344.341 |
4 |
↓
|
|
|
|
Analogs
-
21289021
-
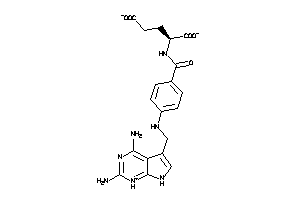
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.10 |
6.64 |
-128.34 |
8 |
12 |
-1 |
216 |
426.413 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.10 |
6.22 |
-120.56 |
7 |
12 |
-2 |
215 |
425.405 |
9 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-2.10 |
4.73 |
-81.56 |
9 |
12 |
0 |
213 |
427.421 |
9 |
↓
|
|
|
|
|
|
|
Analogs
-
36488997
-
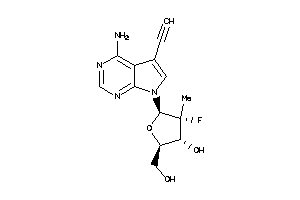
-
36152339
-
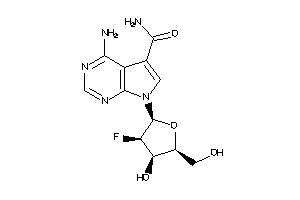
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.21 |
-4.34 |
-15.35 |
6 |
9 |
0 |
150 |
311.273 |
3 |
↓
|
|
|
|
Analogs
-
36152355
-
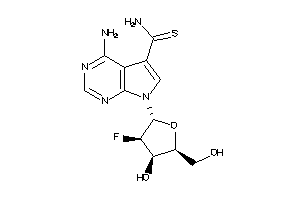
-
27416883
-
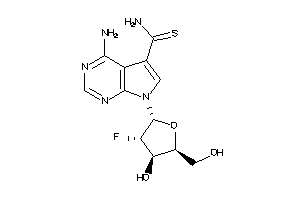
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.23 |
-1.88 |
-18.48 |
6 |
8 |
0 |
132 |
327.341 |
3 |
↓
|
|
|
|
Analogs
-
36152349
-
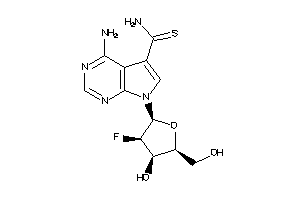
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.23 |
-2.09 |
-16.47 |
6 |
8 |
0 |
132 |
327.341 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.13 |
9.22 |
-15.7 |
2 |
5 |
0 |
81 |
281.294 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.13 |
9.69 |
-33.76 |
3 |
5 |
1 |
82 |
282.302 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.62 |
4.53 |
-18.94 |
2 |
6 |
0 |
91 |
294.38 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.62 |
4.97 |
-38.5 |
3 |
6 |
1 |
92 |
295.388 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.70 |
4.5 |
-19.33 |
2 |
6 |
0 |
91 |
308.407 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.70 |
4.88 |
-35.46 |
3 |
6 |
1 |
92 |
309.415 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.20 |
8.92 |
-10.45 |
2 |
5 |
0 |
60 |
281.363 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.20 |
9.39 |
-25.77 |
3 |
5 |
1 |
61 |
282.371 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.51 |
9.35 |
-11.15 |
2 |
4 |
0 |
57 |
349.207 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.51 |
9.77 |
-23.8 |
3 |
4 |
1 |
58 |
350.215 |
1 |
↓
|
|