|
|
|
|
|
|
|
|
|
|
|
Analogs
-
2010690
-
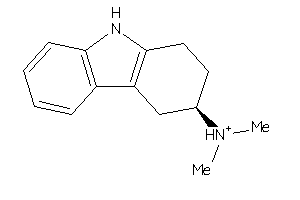
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.83 |
0.67 |
-38.94 |
2 |
2 |
1 |
20 |
215.32 |
1 |
↓
|
|
|
|
Analogs
-
1373
-
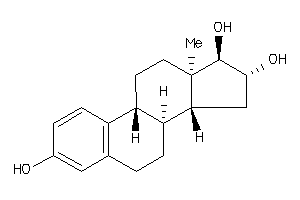
-
8866
-
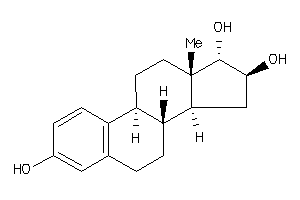
-
525563
-
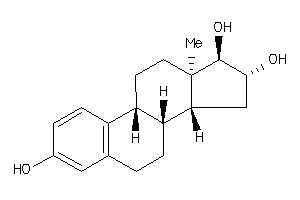
-
525565
-
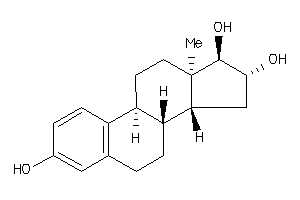
-
525566
-
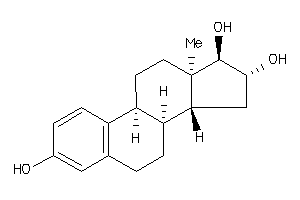
Draw
Identity
99%
90%
80%
70%
Popular Name:
(8S,9R,13R,14R,16S,17R)-13-methyl-6,7,8,9,11,12,14,15,16,17-decahydrocyclopenta[a]phenanthrene-3,16,
(8S,9R,13R,14R,16S,17R)-13-methy…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.51 |
-4.75 |
-6.17 |
3 |
3 |
0 |
60 |
288.387 |
0 |
↓
|
|
|
|
|
|
|
Analogs
-
2018949
-
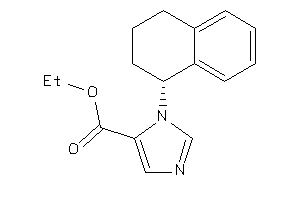
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.80 |
2.73 |
-7.47 |
0 |
4 |
0 |
44 |
270.332 |
4 |
↓
|
|
|
|
Analogs
-
2007119
-
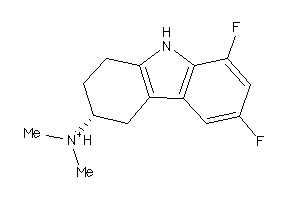
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.09 |
2.16 |
-42.56 |
2 |
2 |
1 |
20 |
251.3 |
1 |
↓
|
|
|
|
Analogs
-
2009618
-
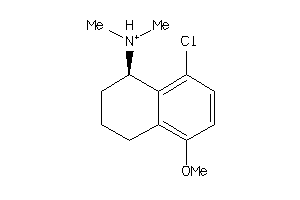
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.34 |
2.4 |
-27.13 |
1 |
2 |
1 |
13 |
240.754 |
2 |
↓
|
|
|
|
|
|
|
Analogs
-
4213852
-
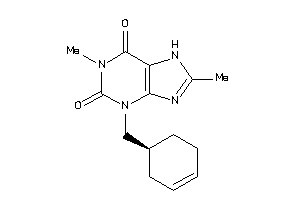
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.88 |
-0.65 |
-9.83 |
1 |
6 |
0 |
72 |
274.324 |
2 |
↓
|
|
|
|
|
|
|
Analogs
-
2555353
-
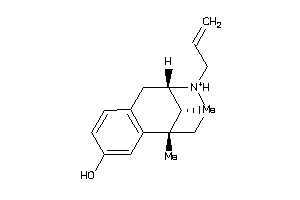
-
2555354
-
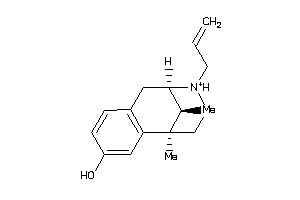
-
7996680
-
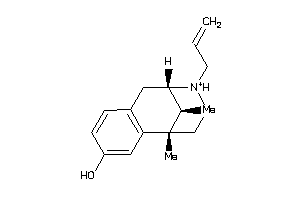
-
13726175
-
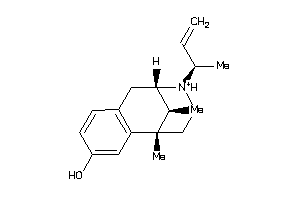
-
13726178
-
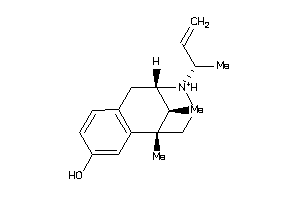
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3B-3-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
504 |
0.46 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
504 |
0.46 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
504 |
0.46 |
Binding ≤ 10μM
|
|
NMDE4-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
504 |
0.46 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.61 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.66 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.62 |
Binding ≤ 10μM
|
|
Z100491-1-O |
Sigma 2 Receptor (cluster #1 Of 2), Other |
Other |
3200 |
0.40 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
5012 |
0.39 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD_MOUSE |
P32300
|
Delta Opioid Receptor, Mouse |
5.5 |
0.61 |
Binding ≤ 1μM
|
|
NMD3B_RAT |
Q8VHN2
|
Glutamate [NMDA] Receptor Subunit 3B, Rat |
504 |
0.46 |
Binding ≤ 1μM
|
|
NMDE2_RAT |
Q00960
|
Glutamate [NMDA] Receptor Subunit Epsilon 2, Rat |
504 |
0.46 |
Binding ≤ 1μM
|
|
NMDE3_RAT |
Q00961
|
Glutamate [NMDA] Receptor Subunit Epsilon 3, Rat |
504 |
0.46 |
Binding ≤ 1μM
|
|
NMDE4_RAT |
Q62645
|
Glutamate [NMDA] Receptor Subunit Epsilon 4, Rat |
504 |
0.46 |
Binding ≤ 1μM
|
|
OPRK_MOUSE |
P33534
|
Kappa Opioid Receptor, Mouse |
0.3 |
0.70 |
Binding ≤ 1μM
|
|
OPRM_MOUSE |
P42866
|
Mu Opioid Receptor, Mouse |
0.9 |
0.67 |
Binding ≤ 1μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
3.6 |
0.62 |
Binding ≤ 1μM
|
|
OPRD_MOUSE |
P32300
|
Delta Opioid Receptor, Mouse |
5.5 |
0.61 |
Binding ≤ 10μM
|
|
NMD3B_RAT |
Q8VHN2
|
Glutamate [NMDA] Receptor Subunit 3B, Rat |
504 |
0.46 |
Binding ≤ 10μM
|
|
NMDE2_RAT |
Q00960
|
Glutamate [NMDA] Receptor Subunit Epsilon 2, Rat |
504 |
0.46 |
Binding ≤ 10μM
|
|
NMDE3_RAT |
Q00961
|
Glutamate [NMDA] Receptor Subunit Epsilon 3, Rat |
504 |
0.46 |
Binding ≤ 10μM
|
|
NMDE4_RAT |
Q62645
|
Glutamate [NMDA] Receptor Subunit Epsilon 4, Rat |
504 |
0.46 |
Binding ≤ 10μM
|
|
OPRK_MOUSE |
P33534
|
Kappa Opioid Receptor, Mouse |
0.3 |
0.70 |
Binding ≤ 10μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
3.6 |
0.62 |
Binding ≤ 10μM
|
|
OPRM_MOUSE |
P42866
|
Mu Opioid Receptor, Mouse |
0.9 |
0.67 |
Binding ≤ 10μM
|
|
Z100491 |
Z100491
|
Sigma 2 Receptor |
3200 |
0.40 |
Binding ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
5011.87234 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.56 |
6.87 |
-36.86 |
2 |
2 |
1 |
25 |
258.385 |
2 |
↓
|
|
|
|
Analogs
-
967790
-
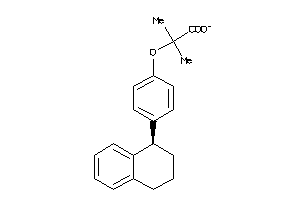
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.39 |
11.98 |
-48.49 |
0 |
3 |
-1 |
49 |
309.385 |
4 |
↓
|
|
|
|
Analogs
-
1846404
-
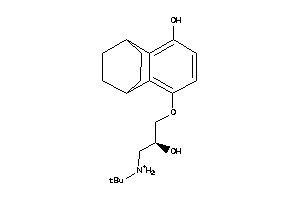
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.91 |
-4.14 |
-41.18 |
4 |
4 |
1 |
66 |
320.453 |
6 |
↓
|
|
|
|
Analogs
-
2008674
-
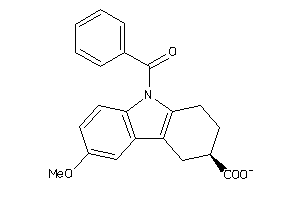
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.01 |
1.88 |
-52.7 |
0 |
5 |
-1 |
71 |
348.378 |
3 |
↓
|
|
|
|
|
|
|
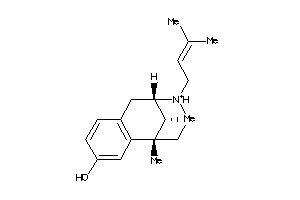
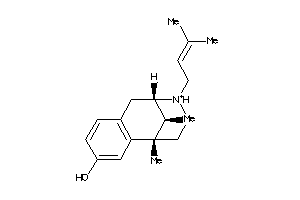
Analogs
-
20242
-
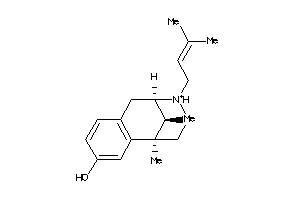
-
1482087
-
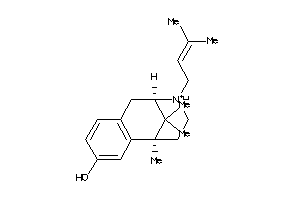
-
1699600
-
-
2015833
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3B-3-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
842 |
0.40 |
Binding ≤ 10μM
|
|
NMDE2-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
842 |
0.40 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
842 |
0.40 |
Binding ≤ 10μM
|
|
NMDE4-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
842 |
0.40 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
49 |
0.49 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
87 |
0.47 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3 |
0.57 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
75 |
0.47 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.54 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
87 |
0.47 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9 |
0.54 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
87 |
0.47 |
Binding ≤ 10μM
|
|
Z100491-1-O |
Sigma 2 Receptor (cluster #1 Of 2), Other |
Other |
29 |
0.50 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD_HUMAN |
P41143
|
Delta Opioid Receptor, Human |
180 |
0.45 |
Binding ≤ 1μM
|
|
OPRD_RAT |
P33533
|
Delta Opioid Receptor, Rat |
12.5 |
0.53 |
Binding ≤ 1μM
|
|
NMD3B_RAT |
Q8VHN2
|
Glutamate [NMDA] Receptor Subunit 3B, Rat |
842 |
0.40 |
Binding ≤ 1μM
|
|
NMDE2_RAT |
Q00960
|
Glutamate [NMDA] Receptor Subunit Epsilon 2, Rat |
842 |
0.40 |
Binding ≤ 1μM
|
|
NMDE3_RAT |
Q00961
|
Glutamate [NMDA] Receptor Subunit Epsilon 3, Rat |
842 |
0.40 |
Binding ≤ 1μM
|
|
NMDE4_RAT |
Q62645
|
Glutamate [NMDA] Receptor Subunit Epsilon 4, Rat |
842 |
0.40 |
Binding ≤ 1μM
|
|
OPRK_HUMAN |
P41145
|
Kappa Opioid Receptor, Human |
75 |
0.47 |
Binding ≤ 1μM
|
|
OPRK_RAT |
P34975
|
Kappa Opioid Receptor, Rat |
12.5 |
0.53 |
Binding ≤ 1μM
|
|
OPRM_HUMAN |
P35372
|
Mu Opioid Receptor, Human |
3.9 |
0.56 |
Binding ≤ 1μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
12.5 |
0.53 |
Binding ≤ 1μM
|
|
Z100491 |
Z100491
|
Sigma 2 Receptor |
29 |
0.50 |
Binding ≤ 1μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
12.5 |
0.53 |
Binding ≤ 1μM
|
|
SGMR1_HUMAN |
Q99720
|
Sigma Opioid Receptor, Human |
3.77 |
0.56 |
Binding ≤ 1μM
|
|
SGMR1_CAVPO |
Q60492
|
Sigma-1 Receptor, Guinea Pig |
3.09 |
0.57 |
Binding ≤ 1μM |
|
OPRD_RAT |
P33533
|
Delta Opioid Receptor, Rat |
12.5 |
0.53 |
Binding ≤ 10μM
|
|
OPRD_HUMAN |
P41143
|
Delta Opioid Receptor, Human |
180 |
0.45 |
Binding ≤ 10μM
|
|
NMD3B_RAT |
Q8VHN2
|
Glutamate [NMDA] Receptor Subunit 3B, Rat |
842 |
0.40 |
Binding ≤ 10μM
|
|
NMDE2_RAT |
Q00960
|
Glutamate [NMDA] Receptor Subunit Epsilon 2, Rat |
842 |
0.40 |
Binding ≤ 10μM
|
|
NMDE3_RAT |
Q00961
|
Glutamate [NMDA] Receptor Subunit Epsilon 3, Rat |
842 |
0.40 |
Binding ≤ 10μM
|
|
NMDE4_RAT |
Q62645
|
Glutamate [NMDA] Receptor Subunit Epsilon 4, Rat |
842 |
0.40 |
Binding ≤ 10μM
|
|
OPRK_RAT |
P34975
|
Kappa Opioid Receptor, Rat |
12.5 |
0.53 |
Binding ≤ 10μM
|
|
OPRK_HUMAN |
P41145
|
Kappa Opioid Receptor, Human |
75 |
0.47 |
Binding ≤ 10μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
12.5 |
0.53 |
Binding ≤ 10μM
|
|
OPRM_HUMAN |
P35372
|
Mu Opioid Receptor, Human |
3.9 |
0.56 |
Binding ≤ 10μM
|
|
Z100491 |
Z100491
|
Sigma 2 Receptor |
29 |
0.50 |
Binding ≤ 10μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
12.5 |
0.53 |
Binding ≤ 10μM
|
|
SGMR1_HUMAN |
Q99720
|
Sigma Opioid Receptor, Human |
3.77 |
0.56 |
Binding ≤ 10μM
|
|
SGMR1_CAVPO |
Q60492
|
Sigma-1 Receptor, Guinea Pig |
3.09 |
0.57 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.59 |
0.88 |
-38.19 |
2 |
2 |
1 |
24 |
286.439 |
2 |
↓
|
|
|
|
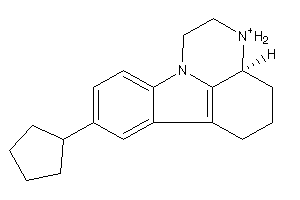
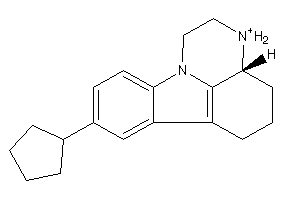
Analogs
-
188861
-
-
188864
-
-
213041
-
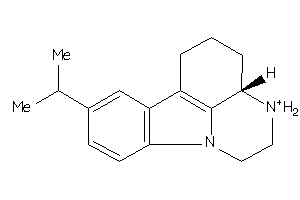
-
213046
-
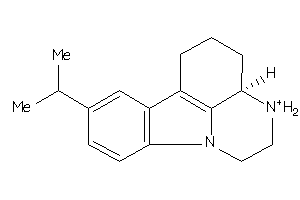
-
281724
-
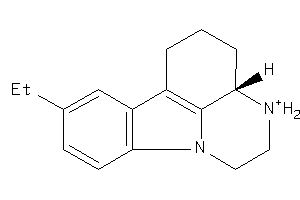
Draw
Identity
99%
90%
80%
70%
Popular Name:
8-Methyl-2,3,3a,4,5,6-hexahydro-1H-pyrazino[3,2,1-jk]carbazole hydrochloride
8-Methyl-2,3,3a,4,5,6-hexahydro-…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.25 |
7.9 |
-47.49 |
2 |
2 |
1 |
22 |
227.331 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.25 |
6.64 |
-6.53 |
1 |
2 |
0 |
17 |
226.323 |
0 |
↓
|
|
|
|
Analogs
-
1843612
-
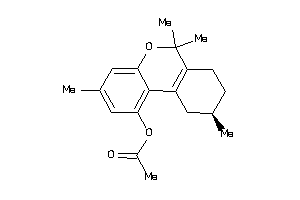
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.11 |
3.42 |
-10.44 |
0 |
3 |
0 |
35 |
300.398 |
2 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
1081249
-
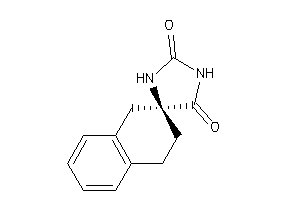
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.30 |
1.99 |
-8.53 |
2 |
4 |
0 |
58 |
216.24 |
0 |
↓
|
|
|
|
Analogs
-
6037100
-
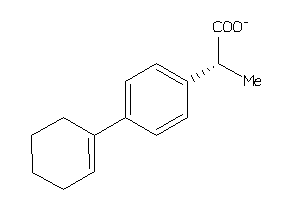
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.04 |
1.69 |
-47.48 |
0 |
2 |
-1 |
40 |
229.299 |
3 |
↓
|
|
|
|
Analogs
-
59299138
-
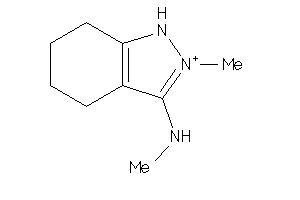
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.54 |
4.21 |
-24.05 |
2 |
3 |
1 |
31 |
166.248 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.54 |
4.09 |
-7.02 |
1 |
3 |
0 |
30 |
165.24 |
1 |
↓
|
|
|
|
Analogs
-
1846110
-
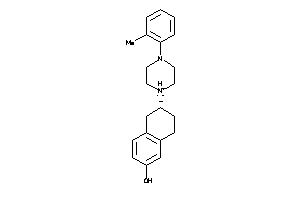
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.13 |
8.95 |
-45.14 |
2 |
3 |
1 |
28 |
323.46 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.13 |
6.69 |
-6.62 |
1 |
3 |
0 |
27 |
322.452 |
2 |
↓
|
|
|
|
Analogs
-
8064059
-
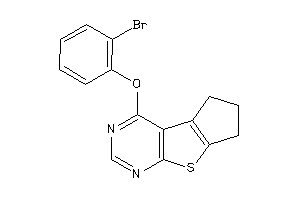
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.16 |
-2.03 |
-5.86 |
0 |
3 |
0 |
35 |
361.264 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.04 |
1 |
-12.68 |
0 |
4 |
0 |
39 |
355.441 |
3 |
↓
|
|
|
|
Analogs
-
2777367
-
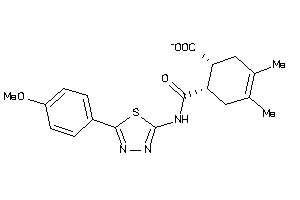
-
2777368
-
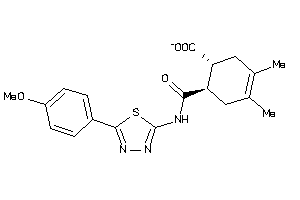
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.66 |
8.22 |
-49.62 |
1 |
6 |
-1 |
95 |
328.373 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.66 |
6.43 |
-16.53 |
2 |
6 |
0 |
92 |
329.381 |
4 |
↓
|
|
|
|
Analogs
-
2777367
-

-
2777368
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.66 |
8.21 |
-66.62 |
1 |
6 |
-1 |
95 |
328.373 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.66 |
7.1 |
-20.5 |
2 |
6 |
0 |
92 |
329.381 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.12 |
-0.59 |
-9.27 |
2 |
6 |
0 |
93 |
310.386 |
1 |
↓
|
|
|
|
Analogs
-
2777367
-

-
2777368
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.66 |
8.37 |
-72.96 |
1 |
6 |
-1 |
95 |
328.373 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.66 |
7.1 |
-20.52 |
2 |
6 |
0 |
92 |
329.381 |
4 |
↓
|
|
|
|
Analogs
-
2777367
-

-
2777368
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.66 |
8.52 |
-51 |
1 |
6 |
-1 |
95 |
328.373 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.66 |
6.43 |
-16.52 |
2 |
6 |
0 |
92 |
329.381 |
4 |
↓
|
|
|
|
Analogs
-
4194956
-
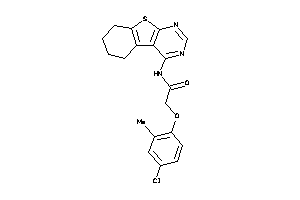
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.74 |
-0.26 |
-16.53 |
1 |
5 |
0 |
64 |
401.919 |
4 |
↓
|
|
|
|
Analogs
-
4194956
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.74 |
-0.33 |
-16.64 |
1 |
5 |
0 |
64 |
401.919 |
4 |
↓
|
|
|
|
Analogs
-
25142043
-
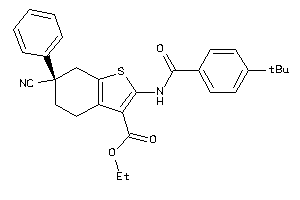
-
25142045
-
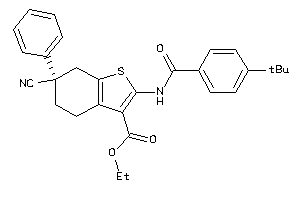
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.10 |
2.46 |
-14.11 |
1 |
5 |
0 |
79 |
430.529 |
6 |
↓
|
|
|
|
Analogs
-
25142043
-

-
25142045
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.10 |
2.46 |
-14.14 |
1 |
5 |
0 |
79 |
430.529 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
2.11 |
-15.78 |
1 |
5 |
0 |
79 |
368.458 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.43 |
2.13 |
-15.74 |
1 |
5 |
0 |
79 |
368.458 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.36 |
2.74 |
-14.56 |
1 |
6 |
0 |
92 |
420.49 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.36 |
2.74 |
-16.07 |
1 |
6 |
0 |
92 |
420.49 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.20 |
2.46 |
-15.12 |
1 |
5 |
0 |
79 |
444.556 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.20 |
2.46 |
-15.11 |
1 |
5 |
0 |
79 |
444.556 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.45 |
7.77 |
-14.52 |
1 |
7 |
0 |
100 |
433.917 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
2.45 |
7.76 |
-13.49 |
1 |
7 |
0 |
100 |
433.917 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
2.45 |
7.73 |
-12.5 |
1 |
7 |
0 |
100 |
433.917 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.74 |
-1.19 |
-19.91 |
1 |
5 |
0 |
64 |
408.31 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.13 |
0.72 |
-11.56 |
1 |
6 |
0 |
91 |
406.463 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.45 |
11 |
-59.89 |
1 |
6 |
-1 |
96 |
372.422 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.74 |
-1.88 |
-13.28 |
1 |
6 |
0 |
64 |
383.521 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.74 |
-1.85 |
-12.89 |
1 |
6 |
0 |
64 |
383.521 |
4 |
↓
|
|