|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.38 |
3.48 |
-12.83 |
3 |
6 |
0 |
83 |
298.346 |
7 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.16 |
8.32 |
-14.54 |
0 |
4 |
0 |
41 |
350.591 |
6 |
↓
|
|
|
|
Analogs
-
21299910
-
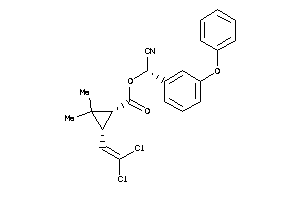
-
607702
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.39 |
14.13 |
-8.57 |
0 |
4 |
0 |
59 |
416.304 |
7 |
↓
|
|
|
|
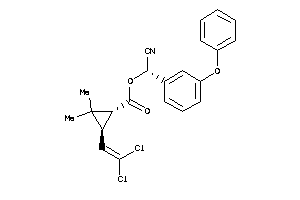
Analogs
-
21299910
-

-
607702
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.39 |
14.2 |
-9.47 |
0 |
4 |
0 |
59 |
416.304 |
7 |
↓
|
|
|
|
Analogs
-
21299910
-

-
607702
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.39 |
14.19 |
-8.81 |
0 |
4 |
0 |
59 |
416.304 |
7 |
↓
|
|
|
|
Analogs
-
21299910
-

-
607702
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.39 |
14.12 |
-8.18 |
0 |
4 |
0 |
59 |
416.304 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.86 |
9.46 |
-14.45 |
0 |
3 |
0 |
28 |
278.335 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.18 |
5.75 |
-16.89 |
2 |
7 |
0 |
93 |
376.416 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.67 |
3.88 |
-11.25 |
1 |
4 |
0 |
48 |
209.245 |
4 |
↓
|
|
|
|
Analogs
-
3798074
-
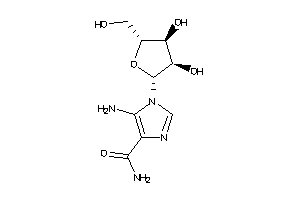
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.50 |
-8.42 |
-19.65 |
7 |
9 |
0 |
157 |
258.234 |
3 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.05 |
5.62 |
-15.26 |
2 |
5 |
0 |
76 |
271.272 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.05 |
6.63 |
-58.56 |
1 |
5 |
-1 |
78 |
270.264 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.56 |
1.3 |
-11.42 |
3 |
2 |
0 |
38 |
116.189 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.48 |
0.61 |
-5.02 |
2 |
2 |
0 |
39 |
100.146 |
0 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.48 |
1.12 |
-27.14 |
3 |
2 |
1 |
40 |
101.154 |
0 |
↓
|
|
|
|
|
|
|
Analogs
-
8214363
-
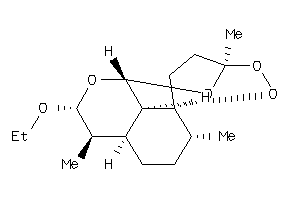
-
14263142
-
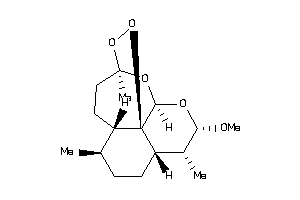
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
4 |
0.53 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.77 |
-2.15 |
-5.89 |
0 |
5 |
0 |
46 |
312.406 |
2 |
↓
|
|
|
|
Analogs
-
14263142
-

-
8214360
-
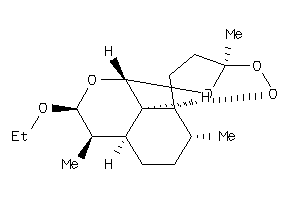
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.77 |
-1.25 |
-5.5 |
0 |
5 |
0 |
46 |
312.406 |
2 |
↓
|
|
|
|
Analogs
-
15449187
-
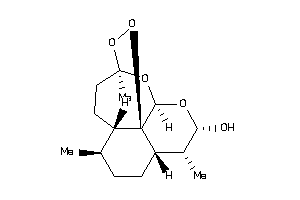
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
56 |
0.51 |
Functional ≤ 10μM
|
|
Z50426-3-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #3 Of 9), Other |
Other |
6 |
0.58 |
Functional ≤ 10μM
|
|
Z50436-3-O |
Filobasidiella Neoformans (cluster #3 Of 3), Other |
Other |
280 |
0.46 |
Functional ≤ 10μM
|
|
Z50442-2-O |
Candida Albicans (cluster #2 Of 4), Other |
Other |
880 |
0.42 |
Functional ≤ 10μM
|
|
Z80156-3-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #3 Of 12), Other |
Other |
2900 |
0.39 |
Functional ≤ 10μM
|
|
Z80482-1-O |
SK-MEL-2 (Melanoma Cells) (cluster #1 Of 4), Other |
Other |
750 |
0.43 |
Functional ≤ 10μM
|
|
Z80493-2-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #2 Of 6), Other |
Other |
850 |
0.42 |
Functional ≤ 10μM
|
|
Z80600-2-O |
XF498 (Glioma Cells) (cluster #2 Of 2), Other |
Other |
540 |
0.44 |
Functional ≤ 10μM
|
|
Z80682-6-O |
A549 (Lung Carcinoma Cells) (cluster #6 Of 11), Other |
Other |
630 |
0.43 |
Functional ≤ 10μM
|
|
Z81057-2-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #2 Of 4), Other |
Other |
8910 |
0.35 |
Functional ≤ 10μM
|
|
Z81072-2-O |
Jurkat (Acute Leukemic T-cells) (cluster #2 Of 10), Other |
Other |
650 |
0.43 |
Functional ≤ 10μM
|
|
Z81335-2-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
460 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.78 |
3.66 |
-6.09 |
1 |
5 |
0 |
57 |
284.352 |
0 |
↓
|
|
|
|
Analogs
-
15449189
-
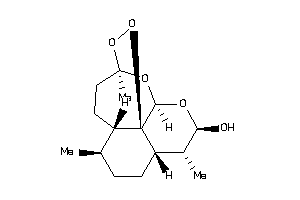
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
56 |
0.51 |
Functional ≤ 10μM
|
|
Z50426-3-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #3 Of 9), Other |
Other |
6 |
0.58 |
Functional ≤ 10μM
|
|
Z50436-3-O |
Filobasidiella Neoformans (cluster #3 Of 3), Other |
Other |
280 |
0.46 |
Functional ≤ 10μM
|
|
Z50442-2-O |
Candida Albicans (cluster #2 Of 4), Other |
Other |
880 |
0.42 |
Functional ≤ 10μM
|
|
Z80156-3-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #3 Of 12), Other |
Other |
2900 |
0.39 |
Functional ≤ 10μM
|
|
Z80482-1-O |
SK-MEL-2 (Melanoma Cells) (cluster #1 Of 4), Other |
Other |
750 |
0.43 |
Functional ≤ 10μM
|
|
Z80493-2-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #2 Of 6), Other |
Other |
850 |
0.42 |
Functional ≤ 10μM
|
|
Z80600-2-O |
XF498 (Glioma Cells) (cluster #2 Of 2), Other |
Other |
540 |
0.44 |
Functional ≤ 10μM
|
|
Z80682-6-O |
A549 (Lung Carcinoma Cells) (cluster #6 Of 11), Other |
Other |
630 |
0.43 |
Functional ≤ 10μM
|
|
Z81057-2-O |
HUVEC (Umbilical Vein Endothelial Cells) (cluster #2 Of 4), Other |
Other |
8910 |
0.35 |
Functional ≤ 10μM
|
|
Z81072-2-O |
Jurkat (Acute Leukemic T-cells) (cluster #2 Of 10), Other |
Other |
650 |
0.43 |
Functional ≤ 10μM
|
|
Z81335-2-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
460 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.78 |
3.29 |
-6.48 |
1 |
5 |
0 |
57 |
284.352 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1-4-E |
Transient Receptor Potential Cation Channel Subfamily A Member 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
319 |
0.91 |
Binding ≤ 10μM |
|
Z80712-4-O |
T47D (Breast Carcinoma Cells) (cluster #4 Of 7), Other |
Other |
7400 |
0.72 |
Functional ≤ 10μM |
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRPA1_HUMAN |
O75762
|
Transient Receptor Potential Cation Channel Subfamily A Member 1, Human |
319 |
0.91 |
Binding ≤ 1μM
|
|
TRPA1_HUMAN |
O75762
|
Transient Receptor Potential Cation Channel Subfamily A Member 1, Human |
319 |
0.91 |
Binding ≤ 10μM
|
|
Z80712 |
Z80712
|
T47D (Breast Carcinoma Cells) |
7400 |
0.72 |
Functional ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.01 |
4.91 |
-2.94 |
0 |
1 |
0 |
12 |
149.218 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.01 |
5.47 |
-29.74 |
1 |
1 |
1 |
14 |
150.226 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80156-12-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #12 Of 12), Other |
Other |
3350 |
0.48 |
Functional ≤ 10μM
|
|
Z80164-6-O |
HT-1080 (Fibrosarcoma Cells) (cluster #6 Of 6), Other |
Other |
900 |
0.53 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.28 |
5.76 |
-10.08 |
0 |
4 |
0 |
53 |
216.192 |
1 |
↓
|
|
|
|
|
|
|
Analogs
-
4800575
-
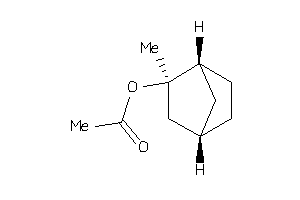
-
4800576
-
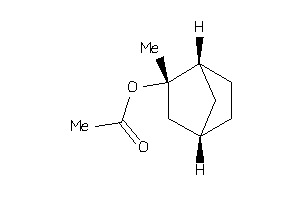
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.05 |
3.24 |
-4.98 |
0 |
2 |
0 |
26 |
196.29 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
3.28 |
-5.93 |
1 |
2 |
0 |
33 |
302.953 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.48 |
4.05 |
-40.83 |
0 |
2 |
-1 |
36 |
301.945 |
0 |
↓
|
|
|
|
Analogs
-
39899474
-
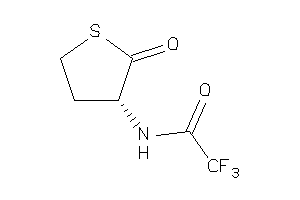
-
39899475
-
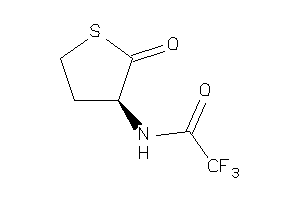
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.02 |
1.29 |
-16.17 |
1 |
3 |
0 |
46 |
159.21 |
1 |
↓
|
|
|
|
Analogs
-
39899474
-

-
39899475
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.02 |
1.29 |
-16.16 |
1 |
3 |
0 |
46 |
159.21 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.20 |
10.96 |
-4.05 |
1 |
1 |
0 |
20 |
365.344 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.44 |
-2.3 |
-10.47 |
3 |
3 |
0 |
63 |
151.165 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.86 |
0.16 |
-11.19 |
2 |
5 |
0 |
70 |
337.46 |
13 |
↓
|
|
|
|
Analogs
-
4212915
-
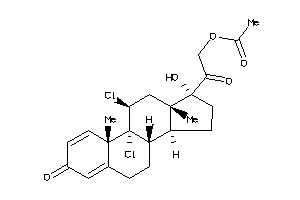
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.83 |
1.43 |
-12.44 |
1 |
5 |
0 |
80 |
455.378 |
4 |
↓
|
|
|
|
Analogs
-
4212915
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.83 |
1.52 |
-12.62 |
1 |
5 |
0 |
80 |
455.378 |
4 |
↓
|
|
|
|
Analogs
-
3875514
-
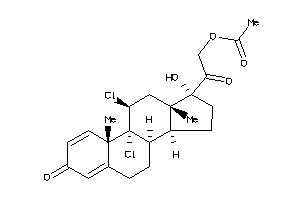
-
3875515
-
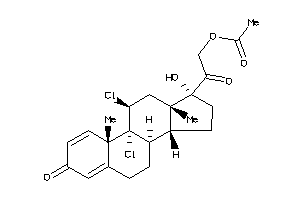
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.83 |
9.39 |
-14 |
1 |
5 |
0 |
81 |
455.378 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.83 |
9.4 |
-13.54 |
1 |
5 |
0 |
81 |
455.378 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-12-E |
Carbonic Anhydrase I (cluster #12 Of 12), Eukaryotic |
Eukaryotes |
2320 |
0.36 |
Binding ≤ 10μM
|
|
CAH14-4-E |
Carbonic Anhydrase XIV (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
8910 |
0.32 |
Binding ≤ 10μM
|
|
CAH2-15-E |
Carbonic Anhydrase II (cluster #15 Of 15), Eukaryotic |
Eukaryotes |
2180 |
0.36 |
Binding ≤ 10μM
|
|
CAH4-14-E |
Carbonic Anhydrase IV (cluster #14 Of 16), Eukaryotic |
Eukaryotes |
9080 |
0.32 |
Binding ≤ 10μM
|
|
CAH5A-6-E |
Carbonic Anhydrase VA (cluster #6 Of 10), Eukaryotic |
Eukaryotes |
7590 |
0.33 |
Binding ≤ 10μM
|
|
CAH6-8-E |
Carbonic Anhydrase VI (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
7060 |
0.33 |
Binding ≤ 10μM
|
|
CAH7-8-E |
Carbonic Anhydrase VII (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
6320 |
0.33 |
Binding ≤ 10μM
|
|
CAH9-11-E |
Carbonic Anhydrase IX (cluster #11 Of 11), Eukaryotic |
Eukaryotes |
9370 |
0.32 |
Binding ≤ 10μM
|
|
CSK21-2-E |
Casein Kinase II Alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
40 |
0.47 |
Binding ≤ 10μM
|
|
CSK2B-3-E |
Casein Kinase II Beta (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
40 |
0.47 |
Binding ≤ 10μM
|
|
ERG1-3-E |
Squalene Monooxygenase (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2000 |
0.36 |
Binding ≤ 10μM
|
|
FGR-2-E |
Tyrosine-protein Kinase FGR (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
9400 |
0.32 |
Binding ≤ 10μM
|
|
GSK3B-7-E |
Glycogen Synthase Kinase-3 Beta (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
7500 |
0.33 |
Binding ≤ 10μM
|
|
KAPCA-3-E |
CAMP-dependent Protein Kinase Alpha-catalytic Subunit (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
600 |
0.40 |
Binding ≤ 10μM
|
|
KAPCB-3-E |
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
600 |
0.40 |
Binding ≤ 10μM
|
|
KAPCG-2-E |
CAMP-dependent Protein Kinase, Gamma Catalytic Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
600 |
0.40 |
Binding ≤ 10μM
|
|
KSYK-2-E |
Tyrosine-protein Kinase SYK (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4300 |
0.34 |
Binding ≤ 10μM
|
|
LYN-1-E |
Tyrosine-protein Kinase Lyn (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2900 |
0.35 |
Binding ≤ 10μM
|
|
SRC-1-E |
Tyrosine-protein Kinase SRC (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
300 |
0.42 |
Binding ≤ 10μM
|
|
Z50136-3-O |
Plasmodium Falciparum (isolate FcB1 / Columbia) (cluster #3 Of 3), Other |
Other |
300 |
0.42 |
Functional ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
330 |
0.41 |
Functional ≤ 10μM
|
|
Z50426-4-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #4 Of 9), Other |
Other |
970 |
0.38 |
Functional ≤ 10μM
|
|
Q7ZJM1-1-V |
Human Immunodeficiency Virus Type 1 Integrase (cluster #1 Of 6), Viral |
Viruses |
5100 |
0.34 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KAPCA_HUMAN |
P17612
|
CAMP-dependent Protein Kinase Alpha-catalytic Subunit, Human |
600 |
0.40 |
Binding ≤ 1μM
|
|
KAPCB_HUMAN |
P22694
|
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit, Human |
600 |
0.40 |
Binding ≤ 1μM
|
|
KAPCG_HUMAN |
P22612
|
CAMP-dependent Protein Kinase, Gamma Catalytic Subunit, Human |
600 |
0.40 |
Binding ≤ 1μM
|
|
CSK21_HUMAN |
P68400
|
Casein Kinase II Alpha, Human |
40 |
0.47 |
Binding ≤ 1μM
|
|
CSK21_RAT |
P19139
|
Casein Kinase II Alpha, Rat |
20 |
0.49 |
Binding ≤ 1μM
|
|
CSK2B_RAT |
P67874
|
Casein Kinase II Beta, Rat |
20 |
0.49 |
Binding ≤ 1μM
|
|
SRC_HUMAN |
P12931
|
Tyrosine-protein Kinase SRC, Human |
300 |
0.42 |
Binding ≤ 1μM
|
|
KAPCA_HUMAN |
P17612
|
CAMP-dependent Protein Kinase Alpha-catalytic Subunit, Human |
3500 |
0.35 |
Binding ≤ 10μM
|
|
KAPCB_HUMAN |
P22694
|
CAMP-dependent Protein Kinase Beta-1 Catalytic Subunit, Human |
3500 |
0.35 |
Binding ≤ 10μM
|
|
KAPCG_HUMAN |
P22612
|
CAMP-dependent Protein Kinase, Gamma Catalytic Subunit, Human |
3500 |
0.35 |
Binding ≤ 10μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
2320 |
0.36 |
Binding ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
2180 |
0.36 |
Binding ≤ 10μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
9080 |
0.32 |
Binding ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
9370 |
0.32 |
Binding ≤ 10μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
7590 |
0.33 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
7060 |
0.33 |
Binding ≤ 10μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
6320 |
0.33 |
Binding ≤ 10μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
8910 |
0.32 |
Binding ≤ 10μM
|
|
CSK21_RAT |
P19139
|
Casein Kinase II Alpha, Rat |
20 |
0.49 |
Binding ≤ 10μM
|
|
CSK21_HUMAN |
P68400
|
Casein Kinase II Alpha, Human |
40 |
0.47 |
Binding ≤ 10μM
|
|
CSK2B_RAT |
P67874
|
Casein Kinase II Beta, Rat |
20 |
0.49 |
Binding ≤ 10μM
|
|
GSK3B_HUMAN |
P49841
|
Glycogen Synthase Kinase-3 Beta, Human |
7500 |
0.33 |
Binding ≤ 10μM
|
|
Q7ZJM1_9HIV1 |
Q7ZJM1
|
Human Immunodeficiency Virus Type 1 Integrase, 9hiv1 |
5100 |
0.34 |
Binding ≤ 10μM
|
|
ERG1_RAT |
P52020
|
Squalene Monooxygenase, Rat |
2000 |
0.36 |
Binding ≤ 10μM
|
|
FGR_RAT |
Q6P6U0
|
Tyrosine-protein Kinase FGR, Rat |
9400 |
0.32 |
Binding ≤ 10μM
|
|
LYN_RAT |
Q07014
|
Tyrosine-protein Kinase Lyn, Rat |
2900 |
0.35 |
Binding ≤ 10μM
|
|
SRC_HUMAN |
P12931
|
Tyrosine-protein Kinase SRC, Human |
300 |
0.42 |
Binding ≤ 10μM
|
|
KSYK_RAT |
Q64725
|
Tyrosine-protein Kinase SYK, Rat |
4300 |
0.34 |
Binding ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
180 |
0.43 |
Functional ≤ 10μM
|
|
Z50136 |
Z50136
|
Plasmodium Falciparum (isolate FcB1 / Columbia) |
300 |
0.42 |
Functional ≤ 10μM
|
|
Z50426 |
Z50426
|
Plasmodium Falciparum (isolate K1 / Thailand) |
970 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.94 |
-1.09 |
-13.29 |
4 |
8 |
0 |
141 |
302.194 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.49 |
-0.46 |
-37.99 |
3 |
8 |
-1 |
144 |
301.186 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.94 |
-0.07 |
-45.06 |
3 |
8 |
-1 |
144 |
301.186 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.02 |
11.11 |
-11.96 |
0 |
5 |
0 |
50 |
395.499 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
5.02 |
11.29 |
-41.15 |
1 |
5 |
1 |
51 |
396.507 |
10 |
↓
|
|
|
|
Analogs
-
608379
-
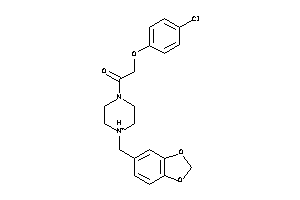
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.20 |
7.74 |
-15.82 |
0 |
6 |
0 |
51 |
388.851 |
5 |
↓
|
|
|
|
Analogs
-
12409058
-
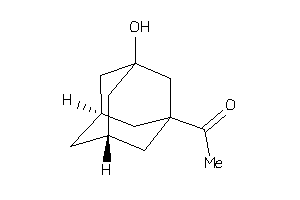
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.14 |
2.39 |
-5.27 |
1 |
2 |
0 |
37 |
166.22 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.31 |
7.44 |
-6.92 |
4 |
5 |
0 |
91 |
256.096 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.07 |
4.68 |
-5.6 |
1 |
3 |
0 |
38 |
137.186 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.29 |
6.27 |
-12.84 |
0 |
5 |
0 |
62 |
260.245 |
2 |
↓
|
|
|
|
Analogs
-
1238584
-
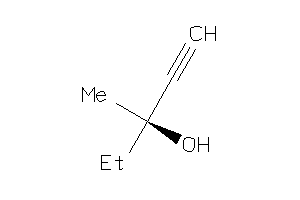
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.15 |
1.44 |
-3.1 |
1 |
1 |
0 |
20 |
98.145 |
1 |
↓
|
|
|
|
Analogs
-
1238583
-
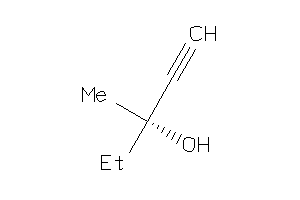
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.15 |
1.4 |
-3.15 |
1 |
1 |
0 |
20 |
98.145 |
1 |
↓
|
|
|
|
Analogs
-
34334111
-
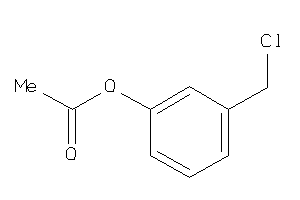
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.91 |
6.38 |
-7.6 |
0 |
2 |
0 |
26 |
150.177 |
2 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.11 |
10.68 |
-12.93 |
1 |
5 |
0 |
73 |
390.52 |
7 |
↓
|
|