|
|
|
|
|
|
|
|
Analogs
-
2555353
-
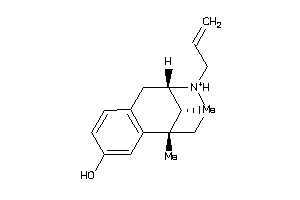
-
2555354
-
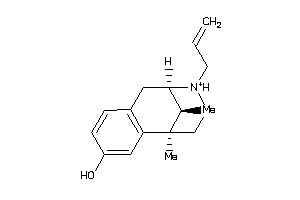
-
7996680
-
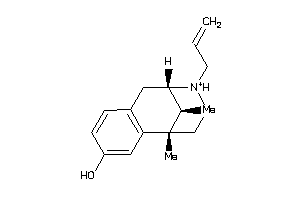
-
13726175
-
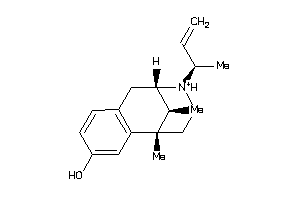
-
13726178
-
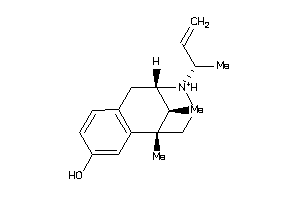
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3B-3-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
504 |
0.46 |
Binding ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
504 |
0.46 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
504 |
0.46 |
Binding ≤ 10μM
|
|
NMDE4-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
504 |
0.46 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.61 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.66 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.62 |
Binding ≤ 10μM
|
|
Z100491-1-O |
Sigma 2 Receptor (cluster #1 Of 2), Other |
Other |
3200 |
0.40 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
5012 |
0.39 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD_MOUSE |
P32300
|
Delta Opioid Receptor, Mouse |
5.5 |
0.61 |
Binding ≤ 1μM
|
|
NMD3B_RAT |
Q8VHN2
|
Glutamate [NMDA] Receptor Subunit 3B, Rat |
504 |
0.46 |
Binding ≤ 1μM
|
|
NMDE2_RAT |
Q00960
|
Glutamate [NMDA] Receptor Subunit Epsilon 2, Rat |
504 |
0.46 |
Binding ≤ 1μM
|
|
NMDE3_RAT |
Q00961
|
Glutamate [NMDA] Receptor Subunit Epsilon 3, Rat |
504 |
0.46 |
Binding ≤ 1μM
|
|
NMDE4_RAT |
Q62645
|
Glutamate [NMDA] Receptor Subunit Epsilon 4, Rat |
504 |
0.46 |
Binding ≤ 1μM
|
|
OPRK_MOUSE |
P33534
|
Kappa Opioid Receptor, Mouse |
0.3 |
0.70 |
Binding ≤ 1μM
|
|
OPRM_MOUSE |
P42866
|
Mu Opioid Receptor, Mouse |
0.9 |
0.67 |
Binding ≤ 1μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
3.6 |
0.62 |
Binding ≤ 1μM
|
|
OPRD_MOUSE |
P32300
|
Delta Opioid Receptor, Mouse |
5.5 |
0.61 |
Binding ≤ 10μM
|
|
NMD3B_RAT |
Q8VHN2
|
Glutamate [NMDA] Receptor Subunit 3B, Rat |
504 |
0.46 |
Binding ≤ 10μM
|
|
NMDE2_RAT |
Q00960
|
Glutamate [NMDA] Receptor Subunit Epsilon 2, Rat |
504 |
0.46 |
Binding ≤ 10μM
|
|
NMDE3_RAT |
Q00961
|
Glutamate [NMDA] Receptor Subunit Epsilon 3, Rat |
504 |
0.46 |
Binding ≤ 10μM
|
|
NMDE4_RAT |
Q62645
|
Glutamate [NMDA] Receptor Subunit Epsilon 4, Rat |
504 |
0.46 |
Binding ≤ 10μM
|
|
OPRK_MOUSE |
P33534
|
Kappa Opioid Receptor, Mouse |
0.3 |
0.70 |
Binding ≤ 10μM
|
|
OPRM_MOUSE |
P42866
|
Mu Opioid Receptor, Mouse |
0.9 |
0.67 |
Binding ≤ 10μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
3.6 |
0.62 |
Binding ≤ 10μM
|
|
Z100491 |
Z100491
|
Sigma 2 Receptor |
3200 |
0.40 |
Binding ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
5011.87234 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.56 |
6.87 |
-36.86 |
2 |
2 |
1 |
25 |
258.385 |
2 |
↓
|
|
|
|
Analogs
-
20242
-
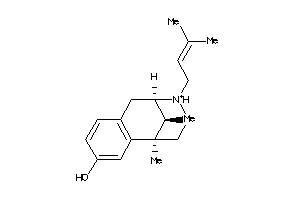
-
1482087
-
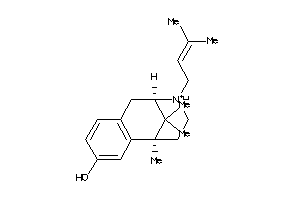
-
1699600
-
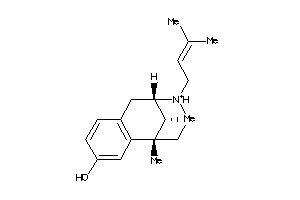
-
2015833
-
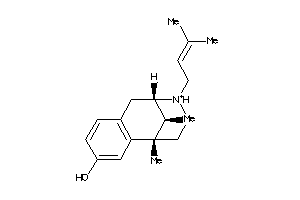
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NMD3B-3-E |
Glutamate [NMDA] Receptor Subunit 3B (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
842 |
0.40 |
Binding ≤ 10μM
|
|
NMDE2-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
842 |
0.40 |
Binding ≤ 10μM
|
|
NMDE3-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
842 |
0.40 |
Binding ≤ 10μM
|
|
NMDE4-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
842 |
0.40 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
49 |
0.49 |
Binding ≤ 10μM
|
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
87 |
0.47 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3 |
0.57 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
75 |
0.47 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
7 |
0.54 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
87 |
0.47 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9 |
0.54 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
87 |
0.47 |
Binding ≤ 10μM
|
|
Z100491-1-O |
Sigma 2 Receptor (cluster #1 Of 2), Other |
Other |
29 |
0.50 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD_HUMAN |
P41143
|
Delta Opioid Receptor, Human |
180 |
0.45 |
Binding ≤ 1μM
|
|
OPRD_RAT |
P33533
|
Delta Opioid Receptor, Rat |
12.5 |
0.53 |
Binding ≤ 1μM
|
|
NMD3B_RAT |
Q8VHN2
|
Glutamate [NMDA] Receptor Subunit 3B, Rat |
842 |
0.40 |
Binding ≤ 1μM
|
|
NMDE2_RAT |
Q00960
|
Glutamate [NMDA] Receptor Subunit Epsilon 2, Rat |
842 |
0.40 |
Binding ≤ 1μM
|
|
NMDE3_RAT |
Q00961
|
Glutamate [NMDA] Receptor Subunit Epsilon 3, Rat |
842 |
0.40 |
Binding ≤ 1μM
|
|
NMDE4_RAT |
Q62645
|
Glutamate [NMDA] Receptor Subunit Epsilon 4, Rat |
842 |
0.40 |
Binding ≤ 1μM
|
|
OPRK_HUMAN |
P41145
|
Kappa Opioid Receptor, Human |
75 |
0.47 |
Binding ≤ 1μM
|
|
OPRK_RAT |
P34975
|
Kappa Opioid Receptor, Rat |
12.5 |
0.53 |
Binding ≤ 1μM
|
|
OPRM_HUMAN |
P35372
|
Mu Opioid Receptor, Human |
3.9 |
0.56 |
Binding ≤ 1μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
12.5 |
0.53 |
Binding ≤ 1μM
|
|
Z100491 |
Z100491
|
Sigma 2 Receptor |
29 |
0.50 |
Binding ≤ 1μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
12.5 |
0.53 |
Binding ≤ 1μM
|
|
SGMR1_HUMAN |
Q99720
|
Sigma Opioid Receptor, Human |
3.77 |
0.56 |
Binding ≤ 1μM
|
|
SGMR1_CAVPO |
Q60492
|
Sigma-1 Receptor, Guinea Pig |
3.09 |
0.57 |
Binding ≤ 1μM |
|
OPRD_HUMAN |
P41143
|
Delta Opioid Receptor, Human |
180 |
0.45 |
Binding ≤ 10μM
|
|
OPRD_RAT |
P33533
|
Delta Opioid Receptor, Rat |
12.5 |
0.53 |
Binding ≤ 10μM
|
|
NMD3B_RAT |
Q8VHN2
|
Glutamate [NMDA] Receptor Subunit 3B, Rat |
842 |
0.40 |
Binding ≤ 10μM
|
|
NMDE2_RAT |
Q00960
|
Glutamate [NMDA] Receptor Subunit Epsilon 2, Rat |
842 |
0.40 |
Binding ≤ 10μM
|
|
NMDE3_RAT |
Q00961
|
Glutamate [NMDA] Receptor Subunit Epsilon 3, Rat |
842 |
0.40 |
Binding ≤ 10μM
|
|
NMDE4_RAT |
Q62645
|
Glutamate [NMDA] Receptor Subunit Epsilon 4, Rat |
842 |
0.40 |
Binding ≤ 10μM
|
|
OPRK_HUMAN |
P41145
|
Kappa Opioid Receptor, Human |
75 |
0.47 |
Binding ≤ 10μM
|
|
OPRK_RAT |
P34975
|
Kappa Opioid Receptor, Rat |
12.5 |
0.53 |
Binding ≤ 10μM
|
|
OPRM_HUMAN |
P35372
|
Mu Opioid Receptor, Human |
3.9 |
0.56 |
Binding ≤ 10μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
12.5 |
0.53 |
Binding ≤ 10μM
|
|
Z100491 |
Z100491
|
Sigma 2 Receptor |
29 |
0.50 |
Binding ≤ 10μM
|
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
12.5 |
0.53 |
Binding ≤ 10μM
|
|
SGMR1_HUMAN |
Q99720
|
Sigma Opioid Receptor, Human |
3.77 |
0.56 |
Binding ≤ 10μM
|
|
SGMR1_CAVPO |
Q60492
|
Sigma-1 Receptor, Guinea Pig |
3.09 |
0.57 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.59 |
0.88 |
-38.19 |
2 |
2 |
1 |
24 |
286.439 |
2 |
↓
|
|
|
|
Analogs
-
13742462
-
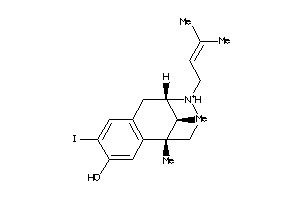
-
13742465
-
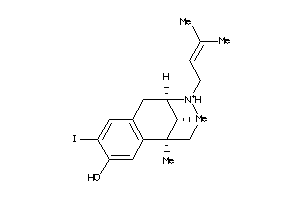
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
94 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.87 |
9.63 |
-37.29 |
2 |
2 |
1 |
25 |
412.335 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2010 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.05 |
10.44 |
-42.68 |
2 |
2 |
1 |
25 |
466.237 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
6.05 |
11.19 |
-58.37 |
1 |
2 |
0 |
27 |
465.229 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
6.05 |
9.34 |
-40.05 |
0 |
2 |
-1 |
26 |
464.221 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2010 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.05 |
10.32 |
-43.72 |
2 |
2 |
1 |
25 |
466.237 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
6.05 |
11.08 |
-60.28 |
1 |
2 |
0 |
27 |
465.229 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
6.05 |
9 |
-40.04 |
0 |
2 |
-1 |
26 |
464.221 |
2 |
↓
|
|
|
|
Analogs
-
13742443
-
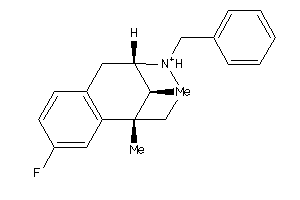
-
13742444
-
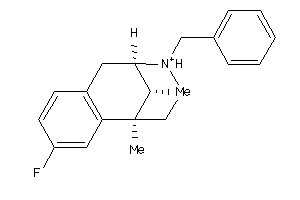
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
43 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.96 |
12.33 |
-40.4 |
1 |
1 |
1 |
4 |
310.436 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.96 |
10.49 |
-3.27 |
0 |
1 |
0 |
3 |
309.428 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1900 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.03 |
8.97 |
-45.61 |
2 |
4 |
1 |
51 |
385.553 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.03 |
7.19 |
-45.6 |
0 |
4 |
-1 |
51 |
383.537 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.03 |
7.12 |
-9.87 |
1 |
4 |
0 |
49 |
384.545 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1900 |
0.30 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.03 |
8.8 |
-46.67 |
2 |
4 |
1 |
51 |
385.553 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.03 |
6.79 |
-45.71 |
0 |
4 |
-1 |
51 |
383.537 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.03 |
6.73 |
-10 |
1 |
4 |
0 |
49 |
384.545 |
4 |
↓
|
|
|
|
Analogs
-
13742439
-
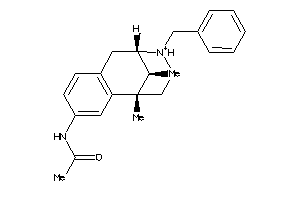
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1410 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.01 |
11.15 |
-42.77 |
2 |
3 |
1 |
34 |
349.498 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.01 |
9.3 |
-9.13 |
1 |
3 |
0 |
32 |
348.49 |
3 |
↓
|
|
|
|
Analogs
-
13742439
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1410 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.01 |
10.99 |
-44.17 |
2 |
3 |
1 |
34 |
349.498 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.01 |
8.89 |
-9.13 |
1 |
3 |
0 |
32 |
348.49 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2037 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.88 |
13.37 |
-39.09 |
1 |
1 |
1 |
4 |
418.342 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.88 |
11.52 |
-2.28 |
0 |
1 |
0 |
3 |
417.334 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2037 |
0.35 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.88 |
13.21 |
-40.2 |
1 |
1 |
1 |
4 |
418.342 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.88 |
11.13 |
-2.39 |
0 |
1 |
0 |
3 |
417.334 |
2 |
↓
|
|
|
|
Analogs
-
13742458
-
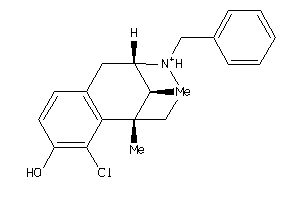
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
764 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.18 |
9.71 |
-41.27 |
2 |
2 |
1 |
25 |
342.89 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.18 |
7.87 |
-4.67 |
1 |
2 |
0 |
23 |
341.882 |
2 |
↓
|
|
|
|
Analogs
-
13742458
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
764 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.18 |
9.62 |
-42.24 |
2 |
2 |
1 |
25 |
342.89 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.18 |
7.54 |
-4.75 |
1 |
2 |
0 |
23 |
341.882 |
2 |
↓
|
|
|
|
Analogs
-
13742463
-
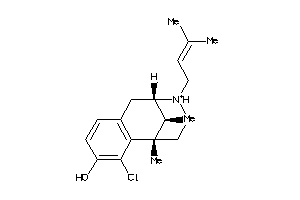
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
4960 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.46 |
8.76 |
-38.22 |
2 |
2 |
1 |
25 |
320.884 |
2 |
↓
|
|
|
|
Analogs
-
13742463
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
4960 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.46 |
8.67 |
-39.28 |
2 |
2 |
1 |
25 |
320.884 |
2 |
↓
|
|
|
|
Analogs
-
34547531
-
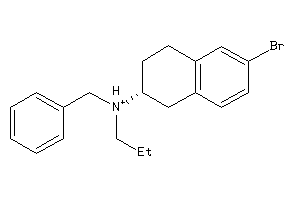
-
34547533
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
501 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.61 |
12.89 |
-39.25 |
1 |
1 |
1 |
4 |
371.342 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.61 |
11.04 |
-2.53 |
0 |
1 |
0 |
3 |
370.334 |
2 |
↓
|
|
|
|
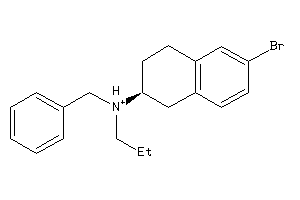
Analogs
-
34547531
-

-
34547533
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
501 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.61 |
12.73 |
-40.52 |
1 |
1 |
1 |
4 |
371.342 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.61 |
10.65 |
-2.59 |
0 |
1 |
0 |
3 |
370.334 |
2 |
↓
|
|
|
|
Analogs
-
13742456
-
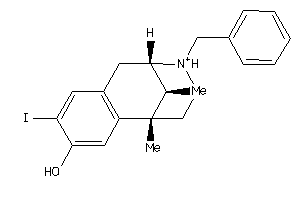
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
597 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.59 |
10.55 |
-39.96 |
2 |
2 |
1 |
25 |
434.341 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.59 |
8.71 |
-3.8 |
1 |
2 |
0 |
23 |
433.333 |
2 |
↓
|
|
|
|
Analogs
-
13742456
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
597 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.59 |
10.39 |
-41.18 |
2 |
2 |
1 |
25 |
434.341 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.59 |
8.31 |
-3.88 |
1 |
2 |
0 |
23 |
433.333 |
2 |
↓
|
|
|
|
Analogs
-
13742460
-
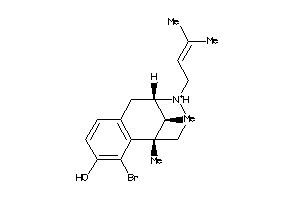
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
283 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.59 |
8.81 |
-38.05 |
2 |
2 |
1 |
25 |
365.335 |
2 |
↓
|
|
|
|
Analogs
-
13742460
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
283 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.59 |
8.7 |
-39.13 |
2 |
2 |
1 |
25 |
365.335 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2170 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.79 |
10.26 |
-43.1 |
2 |
2 |
1 |
25 |
377.335 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.79 |
11.02 |
-61.16 |
1 |
2 |
0 |
27 |
376.327 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.79 |
9.17 |
-41.93 |
0 |
2 |
-1 |
26 |
375.319 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
2170 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.79 |
10.17 |
-44.16 |
2 |
2 |
1 |
25 |
377.335 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.79 |
10.92 |
-63.01 |
1 |
2 |
0 |
27 |
376.327 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.79 |
8.83 |
-41.94 |
0 |
2 |
-1 |
26 |
375.319 |
2 |
↓
|
|
|
|
Analogs
-
13742455
-
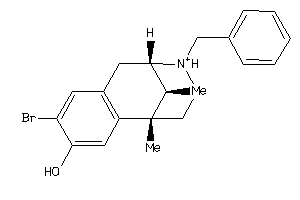
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1220 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.31 |
10.07 |
-40.57 |
2 |
2 |
1 |
25 |
387.341 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.31 |
8.23 |
-4.23 |
1 |
2 |
0 |
23 |
386.333 |
2 |
↓
|
|
|
|
Analogs
-
13742455
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1220 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.31 |
9.91 |
-41.81 |
2 |
2 |
1 |
25 |
387.341 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.31 |
7.84 |
-4.3 |
1 |
2 |
0 |
23 |
386.333 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
211 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.33 |
9.49 |
-39.69 |
2 |
2 |
1 |
25 |
444.231 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
6.33 |
10.25 |
-56.53 |
1 |
2 |
0 |
27 |
443.223 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
211 |
0.41 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.33 |
9.38 |
-40.89 |
2 |
2 |
1 |
25 |
444.231 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
6.33 |
10.13 |
-58.38 |
1 |
2 |
0 |
27 |
443.223 |
2 |
↓
|
|
|
|
Analogs
-
13742454
-
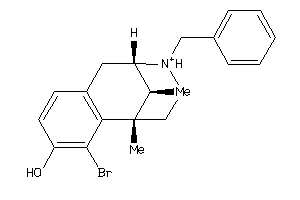
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
346 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.31 |
9.76 |
-41 |
2 |
2 |
1 |
25 |
387.341 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.31 |
7.92 |
-4.6 |
1 |
2 |
0 |
23 |
386.333 |
2 |
↓
|
|
|
|
Analogs
-
13742454
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
346 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.31 |
9.65 |
-42.06 |
2 |
2 |
1 |
25 |
387.341 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.31 |
7.58 |
-4.65 |
1 |
2 |
0 |
23 |
386.333 |
2 |
↓
|
|
|
|
Analogs
-
13742441
-
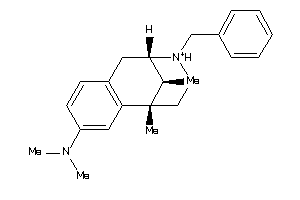
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
258 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.90 |
10.8 |
-3.43 |
0 |
2 |
0 |
6 |
334.507 |
3 |
↓
|
|
|
|
Analogs
-
13742441
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
258 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.90 |
10.41 |
-3.4 |
0 |
2 |
0 |
6 |
334.507 |
3 |
↓
|
|
|
|
Analogs
-
13742451
-
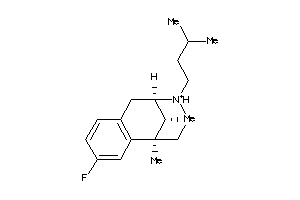
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
56 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.21 |
11.44 |
-38.96 |
1 |
1 |
1 |
4 |
290.446 |
3 |
↓
|
|
|
|
Analogs
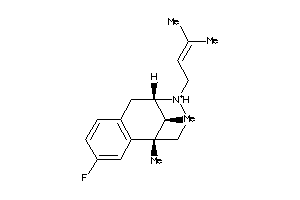
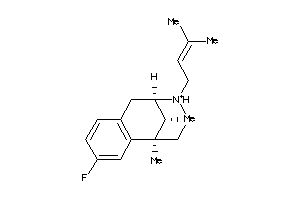
-
13742449
-
-
13742450
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
52 |
0.49 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.24 |
11.41 |
-37.55 |
1 |
1 |
1 |
4 |
288.43 |
2 |
↓
|
|
|
|
Analogs
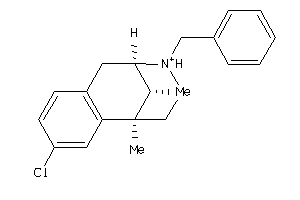
-
13742446
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
429 |
0.39 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.47 |
12.78 |
-39.21 |
1 |
1 |
1 |
4 |
326.891 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.47 |
10.94 |
-2.57 |
0 |
1 |
0 |
3 |
325.883 |
2 |
↓
|
|
|
|
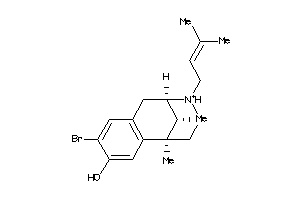
Analogs
-
13742464
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
73 |
0.45 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.59 |
9.15 |
-37.88 |
2 |
2 |
1 |
25 |
365.335 |
2 |
↓
|
|
|
|
Analogs
-
13742440
-
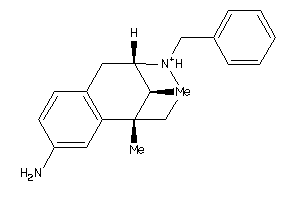
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
832 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.87 |
10.13 |
-34.87 |
3 |
2 |
1 |
30 |
307.461 |
2 |
↓
|
|
|
|
Analogs
-
13742440
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
832 |
0.37 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.87 |
9.96 |
-36.01 |
3 |
2 |
1 |
30 |
307.461 |
2 |
↓
|
|
|
|
Analogs
-
3954257
-
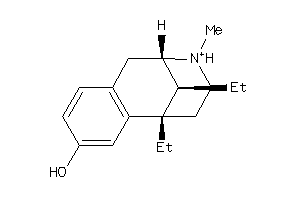
-
4896867
-
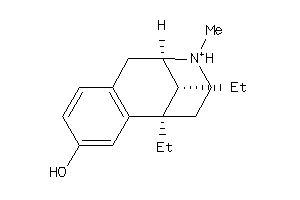
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.87 |
10.17 |
-39.01 |
2 |
2 |
1 |
25 |
316.509 |
6 |
↓
|
|
|
|
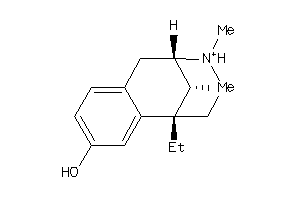
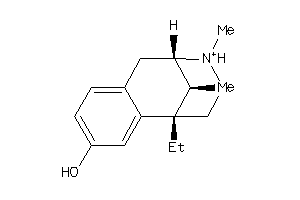
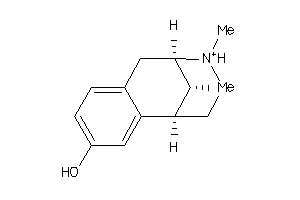
Analogs
-
33824324
-
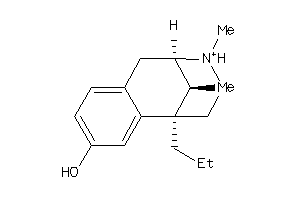
-
31333689
-
-
31333694
-
-
5766169
-
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD_HUMAN |
P41143
|
Delta Opioid Receptor, Human |
57 |
0.44 |
Binding ≤ 1μM
|
|
OPRK_HUMAN |
P41145
|
Kappa Opioid Receptor, Human |
32 |
0.46 |
Binding ≤ 1μM
|
|
OPRM_HUMAN |
P35372
|
Mu Opioid Receptor, Human |
27 |
0.46 |
Binding ≤ 1μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
8.8 |
0.49 |
Binding ≤ 1μM
|
|
SGMR1_HUMAN |
Q99720
|
Sigma Opioid Receptor, Human |
49 |
0.44 |
Binding ≤ 1μM
|
|
OPRD_HUMAN |
P41143
|
Delta Opioid Receptor, Human |
57 |
0.44 |
Binding ≤ 10μM
|
|
Z104302 |
Z104302
|
Glutamate NMDA Receptor |
5634 |
0.32 |
Binding ≤ 10μM
|
|
OPRK_HUMAN |
P41145
|
Kappa Opioid Receptor, Human |
32 |
0.46 |
Binding ≤ 10μM
|
|
OPRM_HUMAN |
P35372
|
Mu Opioid Receptor, Human |
27 |
0.46 |
Binding ≤ 10μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
8.8 |
0.49 |
Binding ≤ 10μM
|
|
SGMR1_HUMAN |
Q99720
|
Sigma Opioid Receptor, Human |
49 |
0.44 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.87 |
10.35 |
-37.93 |
2 |
2 |
1 |
25 |
316.509 |
6 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
33824324
-

-
31333689
-

-
31333694
-

-
5766169
-

Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD_HUMAN |
P41143
|
Delta Opioid Receptor, Human |
464 |
0.35 |
Binding ≤ 1μM
|
|
OPRK_HUMAN |
P41145
|
Kappa Opioid Receptor, Human |
709 |
0.34 |
Binding ≤ 1μM
|
|
OPRM_HUMAN |
P35372
|
Mu Opioid Receptor, Human |
183 |
0.38 |
Binding ≤ 1μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
151 |
0.38 |
Binding ≤ 1μM
|
|
SGMR1_HUMAN |
Q99720
|
Sigma Opioid Receptor, Human |
32 |
0.42 |
Binding ≤ 1μM
|
|
OPRD_HUMAN |
P41143
|
Delta Opioid Receptor, Human |
464 |
0.35 |
Binding ≤ 10μM
|
|
Z104302 |
Z104302
|
Glutamate NMDA Receptor |
6166 |
0.29 |
Binding ≤ 10μM
|
|
OPRK_HUMAN |
P41145
|
Kappa Opioid Receptor, Human |
709 |
0.34 |
Binding ≤ 10μM
|
|
OPRM_HUMAN |
P35372
|
Mu Opioid Receptor, Human |
183 |
0.38 |
Binding ≤ 10μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
151 |
0.38 |
Binding ≤ 10μM
|
|
SGMR1_HUMAN |
Q99720
|
Sigma Opioid Receptor, Human |
32 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.88 |
11.91 |
-37.87 |
2 |
2 |
1 |
25 |
344.563 |
8 |
↓
|
|
|
|
Analogs
-
33824324
-

-
31333689
-

-
31333694
-

-
5766169
-

Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD_HUMAN |
P41143
|
Delta Opioid Receptor, Human |
114 |
0.41 |
Binding ≤ 1μM
|
|
OPRK_HUMAN |
P41145
|
Kappa Opioid Receptor, Human |
77 |
0.41 |
Binding ≤ 1μM
|
|
OPRM_HUMAN |
P35372
|
Mu Opioid Receptor, Human |
59 |
0.42 |
Binding ≤ 1μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
30 |
0.44 |
Binding ≤ 1μM
|
|
SGMR1_HUMAN |
Q99720
|
Sigma Opioid Receptor, Human |
35 |
0.43 |
Binding ≤ 1μM
|
|
OPRD_HUMAN |
P41143
|
Delta Opioid Receptor, Human |
114 |
0.41 |
Binding ≤ 10μM
|
|
Z104302 |
Z104302
|
Glutamate NMDA Receptor |
7975 |
0.30 |
Binding ≤ 10μM
|
|
OPRK_HUMAN |
P41145
|
Kappa Opioid Receptor, Human |
77 |
0.41 |
Binding ≤ 10μM
|
|
OPRM_HUMAN |
P35372
|
Mu Opioid Receptor, Human |
59 |
0.42 |
Binding ≤ 10μM
|
|
OPRM_RAT |
P33535
|
Mu Opioid Receptor, Rat |
30 |
0.44 |
Binding ≤ 10μM
|
|
SGMR1_HUMAN |
Q99720
|
Sigma Opioid Receptor, Human |
35 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.38 |
11.13 |
-37.92 |
2 |
2 |
1 |
25 |
330.536 |
7 |
↓
|
|
|
|
|
|
|
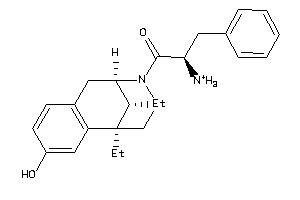
Analogs
-
36215532
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3350 |
0.26 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3350 |
0.26 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3350 |
0.26 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3350 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.73 |
8.17 |
-44.07 |
4 |
4 |
1 |
68 |
393.551 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.73 |
7.88 |
-7.4 |
3 |
4 |
0 |
67 |
392.543 |
5 |
↓
|
|
|
|
Analogs
-
36215530
-
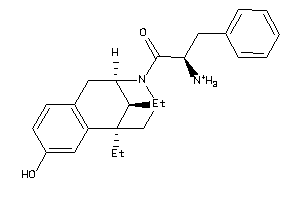
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
OPRD-1-E |
Delta Opioid Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3350 |
0.26 |
Binding ≤ 10μM
|
|
OPRK-1-E |
Kappa Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3350 |
0.26 |
Binding ≤ 10μM
|
|
OPRM-1-E |
Mu-type Opioid Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3350 |
0.26 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
3350 |
0.26 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.73 |
7.78 |
-43.99 |
4 |
4 |
1 |
68 |
393.551 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.73 |
7.49 |
-7.45 |
3 |
4 |
0 |
67 |
392.543 |
5 |
↓
|
|