|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.95 |
4.22 |
-5.89 |
0 |
3 |
0 |
30 |
179.219 |
3 |
↓
|
|
|
|
Analogs
-
7998874
-
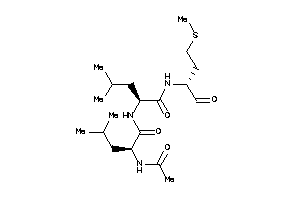
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.55 |
-0.85 |
-17.84 |
3 |
7 |
0 |
104 |
401.573 |
13 |
↓
|
|
|
|
Analogs
-
7998874
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.55 |
-0.85 |
-17.89 |
3 |
7 |
0 |
104 |
401.573 |
13 |
↓
|
|
|
|
Analogs
-
7998874
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.55 |
-1.68 |
-17.54 |
3 |
7 |
0 |
104 |
401.573 |
13 |
↓
|
|
|
|
Analogs
-
7998874
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.55 |
-1.7 |
-19.4 |
3 |
7 |
0 |
104 |
401.573 |
13 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-5-E |
Acetylcholinesterase (cluster #5 Of 12), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
|
CHLE-2-E |
Butyrylcholinesterase (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
7000 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.42 |
12.95 |
-78.82 |
2 |
6 |
2 |
58 |
537.576 |
15 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.23 |
11.67 |
-65.3 |
1 |
6 |
1 |
65 |
536.568 |
15 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.92 |
3.02 |
-11.13 |
3 |
5 |
0 |
93 |
294.31 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE3A_HUMAN |
Q14432
|
Phosphodiesterase 3A, Human |
9100 |
0.35 |
Binding ≤ 10μM
|
|
PDE3B_HUMAN |
Q13370
|
Phosphodiesterase 3B, Human |
9100 |
0.35 |
Binding ≤ 10μM
|
|
PDE4A_RAT |
P54748
|
Phosphodiesterase 4A, Rat |
6000 |
0.37 |
Binding ≤ 10μM
|
|
PDE4A_HUMAN |
P27815
|
Phosphodiesterase 4A, Human |
4800 |
0.37 |
Binding ≤ 10μM
|
|
PDE4B_HUMAN |
Q07343
|
Phosphodiesterase 4B, Human |
1930 |
0.40 |
Binding ≤ 10μM
|
|
PDE4B_RAT |
P14646
|
Phosphodiesterase 4B, Rat |
6000 |
0.37 |
Binding ≤ 10μM
|
|
PDE4C_HUMAN |
Q08493
|
Phosphodiesterase 4C, Human |
4800 |
0.37 |
Binding ≤ 10μM
|
|
PDE4C_RAT |
P14644
|
Phosphodiesterase 4C, Rat |
6000 |
0.37 |
Binding ≤ 10μM
|
|
PDE4D_HUMAN |
Q08499
|
Phosphodiesterase 4D, Human |
4800 |
0.37 |
Binding ≤ 10μM
|
|
PDE4D_RAT |
P14270
|
Phosphodiesterase 4D, Rat |
6000 |
0.37 |
Binding ≤ 10μM
|
|
Z50512 |
Z50512
|
Cavia Porcellus |
120 |
0.48 |
Functional ≤ 10μM
|
|
Z50587 |
Z50587
|
Homo Sapiens |
1800 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.49 |
3.81 |
-14.22 |
2 |
5 |
0 |
60 |
278.352 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PDE3A_HUMAN |
Q14432
|
Phosphodiesterase 3A, Human |
9100 |
0.35 |
Binding ≤ 10μM
|
|
PDE3B_HUMAN |
Q13370
|
Phosphodiesterase 3B, Human |
9100 |
0.35 |
Binding ≤ 10μM
|
|
PDE4A_RAT |
P54748
|
Phosphodiesterase 4A, Rat |
6000 |
0.37 |
Binding ≤ 10μM
|
|
PDE4A_HUMAN |
P27815
|
Phosphodiesterase 4A, Human |
4800 |
0.37 |
Binding ≤ 10μM
|
|
PDE4B_HUMAN |
Q07343
|
Phosphodiesterase 4B, Human |
1930 |
0.40 |
Binding ≤ 10μM
|
|
PDE4B_RAT |
P14646
|
Phosphodiesterase 4B, Rat |
6000 |
0.37 |
Binding ≤ 10μM
|
|
PDE4C_HUMAN |
Q08493
|
Phosphodiesterase 4C, Human |
4800 |
0.37 |
Binding ≤ 10μM
|
|
PDE4C_RAT |
P14644
|
Phosphodiesterase 4C, Rat |
6000 |
0.37 |
Binding ≤ 10μM
|
|
PDE4D_HUMAN |
Q08499
|
Phosphodiesterase 4D, Human |
4800 |
0.37 |
Binding ≤ 10μM
|
|
PDE4D_RAT |
P14270
|
Phosphodiesterase 4D, Rat |
6000 |
0.37 |
Binding ≤ 10μM
|
|
Z50512 |
Z50512
|
Cavia Porcellus |
120 |
0.48 |
Functional ≤ 10μM
|
|
Z50587 |
Z50587
|
Homo Sapiens |
1800 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.49 |
3.82 |
-14.52 |
2 |
5 |
0 |
60 |
278.352 |
7 |
↓
|
|
|
|
Analogs
-
2557944
-
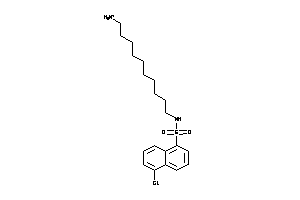
-
1649496
-
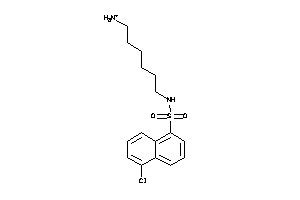
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.75 |
8.16 |
-8.75 |
1 |
3 |
0 |
46 |
339.888 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.18 |
11.6 |
-11.15 |
1 |
3 |
0 |
38 |
331.806 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.40 |
11.19 |
-10.38 |
1 |
3 |
0 |
46 |
401.959 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
10000 |
0.33 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
10000 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.34 |
7.17 |
-10.5 |
1 |
7 |
0 |
81 |
289.339 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A_CHICK |
P49285
|
Melatonin Receptor 1A, Chick |
0.34 |
0.74 |
Binding ≤ 1μM
|
|
MTR1A_HUMAN |
P48039
|
Melatonin Receptor 1A, Human |
0.99 |
0.70 |
Binding ≤ 1μM
|
|
MTR1B_HUMAN |
P49286
|
Melatonin Receptor 1B, Human |
0.99 |
0.70 |
Binding ≤ 1μM
|
|
MTR1B_CHICK |
P51050
|
Melatonin Receptor 1B, Chick |
0.34 |
0.74 |
Binding ≤ 1μM
|
|
MTR1C_CHICK |
P49288
|
Melatonin Receptor 1C, Chick |
0.34 |
0.74 |
Binding ≤ 1μM
|
|
MTR1A_CHICK |
P49285
|
Melatonin Receptor 1A, Chick |
0.34 |
0.74 |
Binding ≤ 10μM
|
|
MTR1A_HUMAN |
P48039
|
Melatonin Receptor 1A, Human |
0.99 |
0.70 |
Binding ≤ 10μM
|
|
MTR1B_CHICK |
P51050
|
Melatonin Receptor 1B, Chick |
0.34 |
0.74 |
Binding ≤ 10μM
|
|
MTR1B_HUMAN |
P49286
|
Melatonin Receptor 1B, Human |
0.99 |
0.70 |
Binding ≤ 10μM
|
|
MTR1C_CHICK |
P49288
|
Melatonin Receptor 1C, Chick |
0.34 |
0.74 |
Binding ≤ 10μM
|
|
MTR1A_HUMAN |
P48039
|
Melatonin Receptor 1A, Human |
0.1 |
0.78 |
Functional ≤ 10μM
|
|
MTR1B_HUMAN |
P49286
|
Melatonin Receptor 1B, Human |
0.1 |
0.78 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.05 |
4.17 |
-14.22 |
2 |
4 |
0 |
54 |
266.728 |
4 |
↓
|
|
|
|
Analogs
-
32131087
-
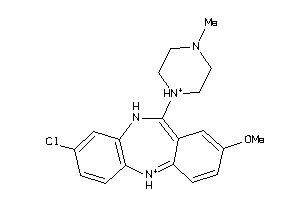
-
32131089
-
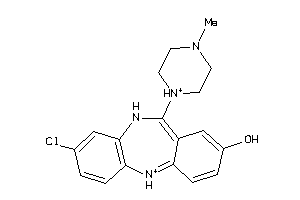
-
3995781
-
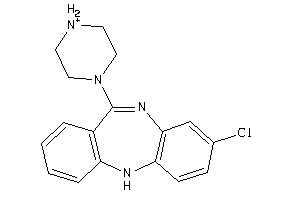
-
4292831
-
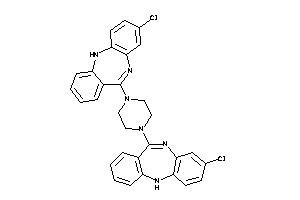
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT1A-4-E |
Serotonin 1a (5-HT1a) Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
2000 |
0.35 |
Binding ≤ 10μM
|
|
5HT1B-1-E |
Serotonin 1b (5-HT1b) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8700 |
0.31 |
Binding ≤ 10μM
|
|
5HT1D-2-E |
Serotonin 1d (5-HT1d) Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
8700 |
0.31 |
Binding ≤ 10μM
|
|
5HT1F-1-E |
Serotonin 1f (5-HT1f) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8700 |
0.31 |
Binding ≤ 10μM
|
|
5HT2A-1-E |
Serotonin 2a (5-HT2a) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
0.52 |
Binding ≤ 10μM
|
|
5HT2B-1-E |
Serotonin 2b (5-HT2b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
28 |
0.46 |
Binding ≤ 10μM
|
|
5HT2C-1-E |
Serotonin 2c (5-HT2c) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
28 |
0.46 |
Binding ≤ 10μM
|
|
5HT3A-1-E |
Serotonin 3a (5-HT3a) Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
53 |
0.44 |
Binding ≤ 10μM
|
|
5HT3B-2-E |
Serotonin 3b (5-HT3b) Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
53 |
0.44 |
Binding ≤ 10μM
|
|
5HT5A-1-E |
Serotonin 5a (5-HT5a) Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.37 |
Binding ≤ 10μM
|
|
5HT6R-1-E |
Serotonin 6 (5-HT6) Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10 |
0.49 |
Binding ≤ 10μM
|
|
5HT7R-1-E |
Serotonin 7 (5-HT7) Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
9 |
0.49 |
Binding ≤ 10μM
|
|
AA3R-1-E |
Adenosine Receptor A3 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
156 |
0.41 |
Binding ≤ 10μM
|
|
ACM1-3-E |
Muscarinic Acetylcholine Receptor M1 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
9 |
0.49 |
Binding ≤ 10μM
|
|
ACM2-4-E |
Muscarinic Acetylcholine Receptor M2 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
91 |
0.43 |
Binding ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
34 |
0.45 |
Binding ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
91 |
0.43 |
Binding ≤ 10μM
|
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
34 |
0.45 |
Binding ≤ 10μM
|
|
ADA1A-1-E |
Alpha-1a Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
ADA1B-1-E |
Alpha-1b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
ADA1D-1-E |
Alpha-1d Adrenergic Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.51 |
Binding ≤ 10μM
|
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
58 |
0.44 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
58 |
0.44 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
58 |
0.44 |
Binding ≤ 10μM
|
|
DRD1-2-E |
Dopamine D1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
8000 |
0.31 |
Binding ≤ 10μM
|
|
DRD3-1-E |
Dopamine D3 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
960 |
0.37 |
Binding ≤ 10μM
|
|
DRD4-4-E |
Dopamine D4 Receptor (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
9 |
0.49 |
Binding ≤ 10μM
|
|
DRD5-1-E |
Dopamine D5 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
890 |
0.37 |
Binding ≤ 10μM
|
|
HRH1-1-E |
Histamine H1 Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2 |
0.53 |
Binding ≤ 10μM
|
|
HRH2-1-E |
Histamine H2 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
18 |
0.47 |
Binding ≤ 10μM
|
|
HRH3-1-E |
Histamine H3 Receptor (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
18 |
0.47 |
Binding ≤ 10μM
|
|
HRH4-1-E |
Histamine H4 Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
70 |
0.44 |
Binding ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
6490 |
0.32 |
Binding ≤ 10μM
|
|
Q63380-1-E |
Transporter (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
390 |
0.39 |
Binding ≤ 10μM
|
|
Q9WTR4-1-E |
Norepinephrine Transporter (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
697 |
0.37 |
Binding ≤ 10μM
|
|
SC6A4-1-E |
Serotonin Transporter (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3900 |
0.33 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
8500 |
0.31 |
Binding ≤ 10μM
|
|
HRH3-2-E |
Histamine H3 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
631 |
0.38 |
Functional ≤ 10μM
|
|
KCNH2-1-E |
HERG (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
191 |
0.41 |
Functional ≤ 10μM
|
|
DRD2-15-E |
Dopamine D2 Receptor (cluster #15 Of 24), Eukaryotic |
Eukaryotes |
290 |
0.40 |
Binding ≤ 10μM
|
|
Z104303-1-O |
Muscarinic Acetylcholine Receptor (cluster #1 Of 7), Other |
Other |
51 |
0.44 |
Binding ≤ 10μM
|
|
Z104304-1-O |
Adrenergic Receptor Alpha-1 (cluster #1 Of 3), Other |
Other |
9 |
0.49 |
Binding ≤ 10μM
|
|
Z102275-1-O |
Hypothalamus (cluster #1 Of 1), Other |
Other |
10 |
0.49 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7943 |
0.31 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
156 |
0.41 |
Binding ≤ 1μM
|
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
1.4 |
0.54 |
Binding ≤ 1μM
|
|
ADA1A_BOVIN |
P18130
|
Alpha-1a Adrenergic Receptor, Bovin |
160 |
0.41 |
Binding ≤ 1μM
|
|
ADA1A_HUMAN |
P35348
|
Alpha-1a Adrenergic Receptor, Human |
11 |
0.48 |
Binding ≤ 1μM
|
|
ADA1A_RAT |
P43140
|
Alpha-1a Adrenergic Receptor, Rat |
17 |
0.47 |
Binding ≤ 1μM
|
|
ADA1B_HUMAN |
P35368
|
Alpha-1b Adrenergic Receptor, Human |
11 |
0.48 |
Binding ≤ 1μM
|
|
ADA1B_RAT |
P15823
|
Alpha-1b Adrenergic Receptor, Rat |
17 |
0.47 |
Binding ≤ 1μM
|
|
ADA1D_HUMAN |
P25100
|
Alpha-1d Adrenergic Receptor, Human |
11 |
0.48 |
Binding ≤ 1μM
|
|
ADA1D_RAT |
P23944
|
Alpha-1d Adrenergic Receptor, Rat |
17 |
0.47 |
Binding ≤ 1μM
|
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
160 |
0.41 |
Binding ≤ 1μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
128 |
0.42 |
Binding ≤ 1μM
|
|
ADA2B_HUMAN |
P18089
|
Alpha-2b Adrenergic Receptor, Human |
160 |
0.41 |
Binding ≤ 1μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
128 |
0.42 |
Binding ≤ 1μM
|
|
ADA2C_HUMAN |
P18825
|
Alpha-2c Adrenergic Receptor, Human |
160 |
0.41 |
Binding ≤ 1μM
|
|
ADA2C_RAT |
P22086
|
Alpha-2c Adrenergic Receptor, Rat |
128 |
0.42 |
Binding ≤ 1μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
114.815362 |
0.42 |
Binding ≤ 1μM
|
|
DRD1_HUMAN |
P21728
|
Dopamine D1 Receptor, Human |
132 |
0.42 |
Binding ≤ 1μM
|
|
DRD1_PIG |
P50130
|
Dopamine D1 Receptor, Pig |
420 |
0.39 |
Binding ≤ 1μM
|
|
DRD1_BOVIN |
Q95136
|
Dopamine D1 Receptor, Bovin |
180 |
0.41 |
Binding ≤ 1μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
1000 |
0.37 |
Binding ≤ 1μM
|
|
DRD2_MOUSE |
P61168
|
Dopamine D2 Receptor, Mouse |
290 |
0.40 |
Binding ≤ 1μM
|
|
DRD2_CHLAE |
P52702
|
Dopamine D2 Receptor, Chlae |
254 |
0.40 |
Binding ≤ 1μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
113 |
0.42 |
Binding ≤ 1μM
|
|
DRD2_BOVIN |
P20288
|
Dopamine D2 Receptor, Bovin |
140 |
0.42 |
Binding ≤ 1μM
|
|
DRD3_RAT |
P19020
|
Dopamine D3 Receptor, Rat |
230 |
0.40 |
Binding ≤ 1μM
|
|
DRD3_HUMAN |
P35462
|
Dopamine D3 Receptor, Human |
180 |
0.41 |
Binding ≤ 1μM
|
|
DRD4_RAT |
P30729
|
Dopamine D4 Receptor, Rat |
247 |
0.40 |
Binding ≤ 1μM
|
|
DRD4_HUMAN |
P21917
|
Dopamine D4 Receptor, Human |
10 |
0.49 |
Binding ≤ 1μM
|
|
DRD5_HUMAN |
P21918
|
Dopamine D5 Receptor, Human |
180 |
0.41 |
Binding ≤ 1μM
|
|
DRD5_RAT |
P25115
|
Dopamine D5 Receptor, Rat |
247 |
0.40 |
Binding ≤ 1μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
190.546072 |
0.41 |
Binding ≤ 1μM
|
|
HRH1_RAT |
P31390
|
Histamine H1 Receptor, Rat |
14 |
0.48 |
Binding ≤ 1μM
|
|
HRH1_HUMAN |
P35367
|
Histamine H1 Receptor, Human |
1.1 |
0.55 |
Binding ≤ 1μM
|
|
HRH2_RAT |
P25102
|
Histamine H2 Receptor, Rat |
18 |
0.47 |
Binding ≤ 1μM
|
|
HRH3_RAT |
Q9QYN8
|
Histamine H3 Receptor, Rat |
18 |
0.47 |
Binding ≤ 1μM
|
|
HRH4_HUMAN |
Q9H3N8
|
Histamine H4 Receptor, Human |
11.9 |
0.48 |
Binding ≤ 1μM
|
|
HRH4_RAT |
Q91ZY1
|
Histamine H4 Receptor, Rat |
18 |
0.47 |
Binding ≤ 1μM
|
|
Z104303 |
Z104303
|
Muscarinic Acetylcholine Receptor |
12 |
0.48 |
Binding ≤ 1μM
|
|
ACM1_RAT |
P08482
|
Muscarinic Acetylcholine Receptor M1, Rat |
2.1 |
0.53 |
Binding ≤ 1μM
|
|
ACM1_HUMAN |
P11229
|
Muscarinic Acetylcholine Receptor M1, Human |
0.98 |
0.55 |
Binding ≤ 1μM
|
|
ACM2_HUMAN |
P08172
|
Muscarinic Acetylcholine Receptor M2, Human |
34 |
0.45 |
Binding ≤ 1μM
|
|
ACM2_RAT |
P10980
|
Muscarinic Acetylcholine Receptor M2, Rat |
46 |
0.45 |
Binding ≤ 1μM
|
|
ACM3_RAT |
P08483
|
Muscarinic Acetylcholine Receptor M3, Rat |
91 |
0.43 |
Binding ≤ 1μM
|
|
ACM3_HUMAN |
P20309
|
Muscarinic Acetylcholine Receptor M3, Human |
34 |
0.45 |
Binding ≤ 1μM
|
|
ACM4_RAT |
P08485
|
Muscarinic Acetylcholine Receptor M4, Rat |
91 |
0.43 |
Binding ≤ 1μM
|
|
ACM4_HUMAN |
P08173
|
Muscarinic Acetylcholine Receptor M4, Human |
34 |
0.45 |
Binding ≤ 1μM
|
|
ACM5_HUMAN |
P08912
|
Muscarinic Acetylcholine Receptor M5, Human |
34 |
0.45 |
Binding ≤ 1μM
|
|
Q9WTR4_RAT |
Q9WTR4
|
Norepinephrine Transporter, Rat |
697 |
0.37 |
Binding ≤ 1μM
|
|
5HT1A_RAT |
P19327
|
Serotonin 1a (5-HT1a) Receptor, Rat |
111 |
0.42 |
Binding ≤ 1μM
|
|
5HT1A_HUMAN |
P08908
|
Serotonin 1a (5-HT1a) Receptor, Human |
140 |
0.42 |
Binding ≤ 1μM
|
|
5HT1B_RAT |
P28564
|
Serotonin 1b (5-HT1b) Receptor, Rat |
210 |
0.41 |
Binding ≤ 1μM
|
|
5HT1D_RAT |
P28565
|
Serotonin 1d (5-HT1d) Receptor, Rat |
210 |
0.41 |
Binding ≤ 1μM
|
|
5HT1F_RAT |
P30940
|
Serotonin 1f (5-HT1f) Receptor, Rat |
210 |
0.41 |
Binding ≤ 1μM
|
|
5HT2A_BOVIN |
Q75Z89
|
Serotonin 2a (5-HT2a) Receptor, Bovin |
3 |
0.52 |
Binding ≤ 1μM
|
|
5HT2A_MOUSE |
P35363
|
Serotonin 2a (5-HT2a) Receptor, Mouse |
28 |
0.46 |
Binding ≤ 1μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
1.8 |
0.53 |
Binding ≤ 1μM
|
|
5HT2A_HUMAN |
P28223
|
Serotonin 2a (5-HT2a) Receptor, Human |
12 |
0.48 |
Binding ≤ 1μM
|
|
5HT2B_HUMAN |
P41595
|
Serotonin 2b (5-HT2b) Receptor, Human |
12 |
0.48 |
Binding ≤ 1μM
|
|
5HT2B_RAT |
P30994
|
Serotonin 2b (5-HT2b) Receptor, Rat |
1.8 |
0.53 |
Binding ≤ 1μM
|
|
5HT2B_MOUSE |
Q02152
|
Serotonin 2b (5-HT2b) Receptor, Mouse |
28 |
0.46 |
Binding ≤ 1μM
|
|
5HT2C_RAT |
P08909
|
Serotonin 2c (5-HT2c) Receptor, Rat |
1.8 |
0.53 |
Binding ≤ 1μM
|
|
5HT2C_HUMAN |
P28335
|
Serotonin 2c (5-HT2c) Receptor, Human |
10 |
0.49 |
Binding ≤ 1μM
|
|
5HT2C_MOUSE |
P34968
|
Serotonin 2c (5-HT2c) Receptor, Mouse |
28 |
0.46 |
Binding ≤ 1μM
|
|
5HT3A_MOUSE |
P23979
|
Serotonin 3a (5-HT3a) Receptor, Mouse |
32 |
0.46 |
Binding ≤ 1μM
|
|
5HT3A_RAT |
P35563
|
Serotonin 3a (5-HT3a) Receptor, Rat |
53 |
0.44 |
Binding ≤ 1μM
|
|
5HT3B_RAT |
Q9JJ16
|
Serotonin 3b (5-HT3b) Receptor, Rat |
53 |
0.44 |
Binding ≤ 1μM
|
|
5HT5A_HUMAN |
P47898
|
Serotonin 5a (5-HT5a) Receptor, Human |
1000 |
0.37 |
Binding ≤ 1μM
|
|
5HT6R_HUMAN |
P50406
|
Serotonin 6 (5-HT6) Receptor, Human |
10 |
0.49 |
Binding ≤ 1μM
|
|
5HT7R_HUMAN |
P34969
|
Serotonin 7 (5-HT7) Receptor, Human |
39.8107171 |
0.45 |
Binding ≤ 1μM
|
|
5HT7R_RAT |
P32305
|
Serotonin 7 (5-HT7) Receptor, Rat |
6.3 |
0.50 |
Binding ≤ 1μM
|
|
Q63380_RAT |
Q63380
|
Transporter, Rat |
390 |
0.39 |
Binding ≤ 1μM
|
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
156 |
0.41 |
Binding ≤ 10μM
|
|
Z104304 |
Z104304
|
Adrenergic Receptor Alpha-1 |
1.4 |
0.54 |
Binding ≤ 10μM
|
|
ADA1A_BOVIN |
P18130
|
Alpha-1a Adrenergic Receptor, Bovin |
160 |
0.41 |
Binding ≤ 10μM
|
|
ADA1A_HUMAN |
P35348
|
Alpha-1a Adrenergic Receptor, Human |
11 |
0.48 |
Binding ≤ 10μM
|
|
ADA1A_RAT |
P43140
|
Alpha-1a Adrenergic Receptor, Rat |
17 |
0.47 |
Binding ≤ 10μM
|
|
ADA1B_HUMAN |
P35368
|
Alpha-1b Adrenergic Receptor, Human |
11 |
0.48 |
Binding ≤ 10μM
|
|
ADA1B_RAT |
P15823
|
Alpha-1b Adrenergic Receptor, Rat |
17 |
0.47 |
Binding ≤ 10μM
|
|
ADA1D_RAT |
P23944
|
Alpha-1d Adrenergic Receptor, Rat |
17 |
0.47 |
Binding ≤ 10μM
|
|
ADA1D_HUMAN |
P25100
|
Alpha-1d Adrenergic Receptor, Human |
11 |
0.48 |
Binding ≤ 10μM
|
|
ADA2A_HUMAN |
P08913
|
Alpha-2a Adrenergic Receptor, Human |
160 |
0.41 |
Binding ≤ 10μM
|
|
ADA2A_RAT |
P22909
|
Alpha-2a Adrenergic Receptor, Rat |
128 |
0.42 |
Binding ≤ 10μM
|
|
ADA2B_HUMAN |
P18089
|
Alpha-2b Adrenergic Receptor, Human |
160 |
0.41 |
Binding ≤ 10μM
|
|
ADA2B_RAT |
P19328
|
Alpha-2b Adrenergic Receptor, Rat |
128 |
0.42 |
Binding ≤ 10μM
|
|
ADA2C_HUMAN |
P18825
|
Alpha-2c Adrenergic Receptor, Human |
160 |
0.41 |
Binding ≤ 10μM
|
|
ADA2C_RAT |
P22086
|
Alpha-2c Adrenergic Receptor, Rat |
128 |
0.42 |
Binding ≤ 10μM
|
|
DRD1_RAT |
P18901
|
Dopamine D1 Receptor, Rat |
114.815362 |
0.42 |
Binding ≤ 10μM
|
|
DRD1_BOVIN |
Q95136
|
Dopamine D1 Receptor, Bovin |
180 |
0.41 |
Binding ≤ 10μM
|
|
DRD1_HUMAN |
P21728
|
Dopamine D1 Receptor, Human |
132 |
0.42 |
Binding ≤ 10μM
|
|
DRD1_PIG |
P50130
|
Dopamine D1 Receptor, Pig |
420 |
0.39 |
Binding ≤ 10μM
|
|
DRD2_RAT |
P61169
|
Dopamine D2 Receptor, Rat |
1000 |
0.37 |
Binding ≤ 10μM
|
|
DRD2_MOUSE |
P61168
|
Dopamine D2 Receptor, Mouse |
290 |
0.40 |
Binding ≤ 10μM
|
|
DRD2_CHLAE |
P52702
|
Dopamine D2 Receptor, Chlae |
254 |
0.40 |
Binding ≤ 10μM
|
|
DRD2_HUMAN |
P14416
|
Dopamine D2 Receptor, Human |
113 |
0.42 |
Binding ≤ 10μM
|
|
DRD2_BOVIN |
P20288
|
Dopamine D2 Receptor, Bovin |
140 |
0.42 |
Binding ≤ 10μM
|
|
DRD3_HUMAN |
P35462
|
Dopamine D3 Receptor, Human |
180 |
0.41 |
Binding ≤ 10μM
|
|
DRD3_RAT |
P19020
|
Dopamine D3 Receptor, Rat |
230 |
0.40 |
Binding ≤ 10μM
|
|
DRD4_RAT |
P30729
|
Dopamine D4 Receptor, Rat |
247 |
0.40 |
Binding ≤ 10μM
|
|
DRD4_HUMAN |
P21917
|
Dopamine D4 Receptor, Human |
10 |
0.49 |
Binding ≤ 10μM
|
|
DRD5_HUMAN |
P21918
|
Dopamine D5 Receptor, Human |
180 |
0.41 |
Binding ≤ 10μM
|
|
DRD5_RAT |
P25115
|
Dopamine D5 Receptor, Rat |
247 |
0.40 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
190.546072 |
0.41 |
Binding ≤ 10μM
|
|
HRH1_RAT |
P31390
|
Histamine H1 Receptor, Rat |
14 |
0.48 |
Binding ≤ 10μM
|
|
HRH1_HUMAN |
P35367
|
Histamine H1 Receptor, Human |
1.1 |
0.55 |
Binding ≤ 10μM
|
|
HRH2_RAT |
P25102
|
Histamine H2 Receptor, Rat |
18 |
0.47 |
Binding ≤ 10μM
|
|
HRH3_RAT |
Q9QYN8
|
Histamine H3 Receptor, Rat |
18 |
0.47 |
Binding ≤ 10μM
|
|
HRH4_HUMAN |
Q9H3N8
|
Histamine H4 Receptor, Human |
11.9 |
0.48 |
Binding ≤ 10μM
|
|
HRH4_RAT |
Q91ZY1
|
Histamine H4 Receptor, Rat |
18 |
0.47 |
Binding ≤ 10μM
|
|
Z104303 |
Z104303
|
Muscarinic Acetylcholine Receptor |
12 |
0.48 |
Binding ≤ 10μM
|
|
ACM1_RAT |
P08482
|
Muscarinic Acetylcholine Receptor M1, Rat |
2.1 |
0.53 |
Binding ≤ 10μM
|
|
ACM1_HUMAN |
P11229
|
Muscarinic Acetylcholine Receptor M1, Human |
0.98 |
0.55 |
Binding ≤ 10μM
|
|
ACM2_HUMAN |
P08172
|
Muscarinic Acetylcholine Receptor M2, Human |
34 |
0.45 |
Binding ≤ 10μM
|
|
ACM2_RAT |
P10980
|
Muscarinic Acetylcholine Receptor M2, Rat |
46 |
0.45 |
Binding ≤ 10μM
|
|
ACM3_RAT |
P08483
|
Muscarinic Acetylcholine Receptor M3, Rat |
91 |
0.43 |
Binding ≤ 10μM
|
|
ACM3_HUMAN |
P20309
|
Muscarinic Acetylcholine Receptor M3, Human |
34 |
0.45 |
Binding ≤ 10μM
|
|
ACM4_RAT |
P08485
|
Muscarinic Acetylcholine Receptor M4, Rat |
91 |
0.43 |
Binding ≤ 10μM
|
|
ACM4_HUMAN |
P08173
|
Muscarinic Acetylcholine Receptor M4, Human |
34 |
0.45 |
Binding ≤ 10μM
|
|
ACM5_HUMAN |
P08912
|
Muscarinic Acetylcholine Receptor M5, Human |
34 |
0.45 |
Binding ≤ 10μM
|
|
Q9WTR4_RAT |
Q9WTR4
|
Norepinephrine Transporter, Rat |
697 |
0.37 |
Binding ≤ 10μM
|
|
5HT1A_HUMAN |
P08908
|
Serotonin 1a (5-HT1a) Receptor, Human |
140 |
0.42 |
Binding ≤ 10μM
|
|
5HT1A_MOUSE |
Q64264
|
Serotonin 1a (5-HT1a) Receptor, Mouse |
2000 |
0.35 |
Binding ≤ 10μM
|
|
5HT1A_RAT |
P19327
|
Serotonin 1a (5-HT1a) Receptor, Rat |
1010 |
0.36 |
Binding ≤ 10μM
|
|
5HT1B_RAT |
P28564
|
Serotonin 1b (5-HT1b) Receptor, Rat |
210 |
0.41 |
Binding ≤ 10μM
|
|
5HT1D_RAT |
P28565
|
Serotonin 1d (5-HT1d) Receptor, Rat |
210 |
0.41 |
Binding ≤ 10μM
|
|
5HT1F_RAT |
P30940
|
Serotonin 1f (5-HT1f) Receptor, Rat |
210 |
0.41 |
Binding ≤ 10μM
|
|
5HT2A_MOUSE |
P35363
|
Serotonin 2a (5-HT2a) Receptor, Mouse |
28 |
0.46 |
Binding ≤ 10μM
|
|
5HT2A_RAT |
P14842
|
Serotonin 2a (5-HT2a) Receptor, Rat |
1.8 |
0.53 |
Binding ≤ 10μM
|
|
5HT2A_HUMAN |
P28223
|
Serotonin 2a (5-HT2a) Receptor, Human |
12 |
0.48 |
Binding ≤ 10μM
|
|
5HT2A_BOVIN |
Q75Z89
|
Serotonin 2a (5-HT2a) Receptor, Bovin |
3 |
0.52 |
Binding ≤ 10μM
|
|
5HT2B_RAT |
P30994
|
Serotonin 2b (5-HT2b) Receptor, Rat |
1.8 |
0.53 |
Binding ≤ 10μM
|
|
5HT2B_MOUSE |
Q02152
|
Serotonin 2b (5-HT2b) Receptor, Mouse |
28 |
0.46 |
Binding ≤ 10μM
|
|
5HT2B_HUMAN |
P41595
|
Serotonin 2b (5-HT2b) Receptor, Human |
12 |
0.48 |
Binding ≤ 10μM
|
|
5HT2C_RAT |
P08909
|
Serotonin 2c (5-HT2c) Receptor, Rat |
1.8 |
0.53 |
Binding ≤ 10μM
|
|
5HT2C_HUMAN |
P28335
|
Serotonin 2c (5-HT2c) Receptor, Human |
10 |
0.49 |
Binding ≤ 10μM
|
|
5HT2C_MOUSE |
P34968
|
Serotonin 2c (5-HT2c) Receptor, Mouse |
28 |
0.46 |
Binding ≤ 10μM
|
|
5HT3A_MOUSE |
P23979
|
Serotonin 3a (5-HT3a) Receptor, Mouse |
32 |
0.46 |
Binding ≤ 10μM
|
|
5HT3A_RAT |
P35563
|
Serotonin 3a (5-HT3a) Receptor, Rat |
53 |
0.44 |
Binding ≤ 10μM
|
|
5HT3B_RAT |
Q9JJ16
|
Serotonin 3b (5-HT3b) Receptor, Rat |
53 |
0.44 |
Binding ≤ 10μM
|
|
5HT5A_HUMAN |
P47898
|
Serotonin 5a (5-HT5a) Receptor, Human |
1000 |
0.37 |
Binding ≤ 10μM
|
|
5HT6R_HUMAN |
P50406
|
Serotonin 6 (5-HT6) Receptor, Human |
10 |
0.49 |
Binding ≤ 10μM
|
|
5HT7R_HUMAN |
P34969
|
Serotonin 7 (5-HT7) Receptor, Human |
39.8107171 |
0.45 |
Binding ≤ 10μM
|
|
5HT7R_RAT |
P32305
|
Serotonin 7 (5-HT7) Receptor, Rat |
6.3 |
0.50 |
Binding ≤ 10μM
|
|
SC6A4_HUMAN |
P31645
|
Serotonin Transporter, Human |
3900 |
0.33 |
Binding ≤ 10μM
|
|
SGMR1_HUMAN |
Q99720
|
Sigma Opioid Receptor, Human |
8500 |
0.31 |
Binding ≤ 10μM
|
|
Q63380_RAT |
Q63380
|
Transporter, Rat |
390 |
0.39 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
191 |
0.41 |
Functional ≤ 10μM
|
|
HRH3_HUMAN |
Q9Y5N1
|
Histamine H3 Receptor, Human |
631 |
0.38 |
Functional ≤ 10μM
|
|
Z102275 |
Z102275
|
Hypothalamus |
10 |
0.49 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
3981.07171 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.14 |
7.55 |
-7.08 |
1 |
4 |
0 |
35 |
326.831 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.14 |
8.88 |
-23.43 |
2 |
4 |
1 |
36 |
327.839 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.14 |
11.23 |
-89.99 |
3 |
4 |
2 |
38 |
328.847 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAN1-1-E |
Calpain 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
7 |
0.44 |
Binding ≤ 10μM
|
|
CATK-1-E |
Cathepsin K (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.84 |
8.3 |
-11.2 |
2 |
6 |
0 |
85 |
362.47 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.44 |
3.11 |
-13.33 |
0 |
4 |
0 |
54 |
273.741 |
1 |
↓
|
|
|
|
Analogs
-
4468952
-
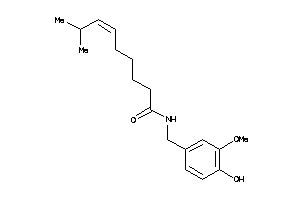
-
33845128
-
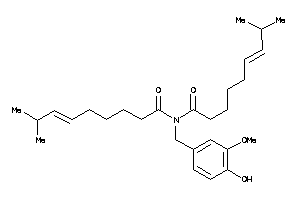
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.10 |
6.6 |
-9.89 |
2 |
4 |
0 |
59 |
305.418 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.93 |
7.74 |
-18.05 |
3 |
4 |
0 |
56 |
376.909 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.58 |
-2.83 |
-15.15 |
5 |
5 |
0 |
98 |
217.14 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.59 |
9.91 |
-5.64 |
1 |
3 |
0 |
48 |
282.387 |
3 |
↓
|
|
|
|
Analogs
-
532225
-
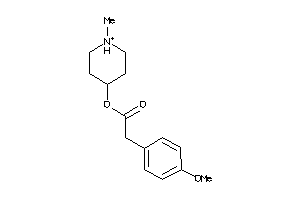
-
6777
-
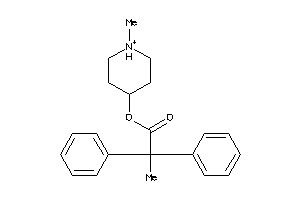
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA3R-3-E |
Adenosine Receptor A3 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
2 |
0.51 |
Binding ≤ 10μM
|
|
ACM3-4-E |
Muscarinic Acetylcholine Receptor M3 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
67 |
0.42 |
Binding ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
1 |
0.53 |
Binding ≤ 10μM
|
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
1 |
0.53 |
Binding ≤ 10μM
|
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
3981 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.19 |
14.24 |
-35.8 |
0 |
3 |
1 |
26 |
324.444 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.15 |
13.51 |
-7.64 |
0 |
4 |
0 |
53 |
314.466 |
17 |
↓
|
|
|
|
Analogs
-
34541434
-
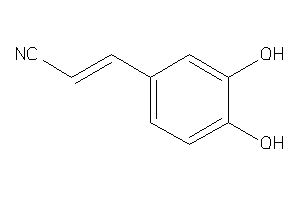
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.20 |
1.39 |
-15.47 |
2 |
4 |
0 |
88 |
186.17 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.20 |
2.31 |
-47.54 |
1 |
4 |
-1 |
91 |
185.162 |
1 |
↓
|
|
|
|
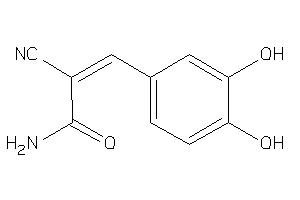
Analogs
-
160657
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4500 |
0.50 |
Binding ≤ 10μM
|
|
ERBB2-1-E |
Receptor Protein-tyrosine Kinase ErbB-2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6400 |
0.48 |
Binding ≤ 10μM
|
|
LCK-3-E |
Tyrosine-protein Kinase LCK (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
7000 |
0.48 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.15 |
-2.32 |
-12.35 |
4 |
5 |
0 |
107 |
204.185 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM2-3-E |
Muscarinic Acetylcholine Receptor M2 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
1440 |
0.20 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM2_PIG |
P06199
|
Muscarinic Acetylcholine Receptor M2, Pig |
1440 |
0.20 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.06 |
5.89 |
-86.97 |
0 |
8 |
2 |
78 |
548.728 |
15 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1-6-E |
GABA Receptor Alpha-1 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRA2-5-E |
GABA Receptor Alpha-2 Subunit (cluster #5 Of 8), Eukaryotic |
Eukaryotes |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRA3-7-E |
GABA Receptor Alpha-3 Subunit (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRA5-3-E |
GABA Receptor Alpha-5 Subunit (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRA6-8-E |
GABA Receptor Alpha-6 Subunit (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRB1-1-E |
GABA Receptor Beta-1 Subunit (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRB2-6-E |
GABA Receptor Beta-2 Subunit (cluster #6 Of 7), Eukaryotic |
Eukaryotes |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRB3-2-E |
GABA Receptor Beta-3 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRD-5-E |
GABA Receptor Delta Subunit (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRE-5-E |
GABA Receptor Epsilon Subunit (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRG1-1-E |
GABA Receptor Gamma-1 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRG2-2-E |
GABA Receptor Gamma-2 Subunit (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRP-5-E |
GABA Receptor Pi Subunit (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRR1-1-E |
GABA Receptor Rho-1 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRR2-1-E |
GABA Receptor Rho-2 Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
197 |
0.55 |
Binding ≤ 10μM
|
|
Q91ZM7-1-E |
GABA Receptor Theta Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
197 |
0.55 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
GBRA1_MOUSE |
P62812
|
GABA Receptor Alpha-1 Subunit, Mouse |
197 |
0.55 |
Binding ≤ 1μM
|
|
GBRA2_RAT |
P23576
|
GABA Receptor Alpha-2 Subunit, Rat |
197 |
0.55 |
Binding ≤ 1μM
|
|
GBRA3_RAT |
P20236
|
GABA Receptor Alpha-3 Subunit, Rat |
197 |
0.55 |
Binding ≤ 1μM
|
|
GBRA4_RAT |
P28471
|
GABA Receptor Alpha-4 Subunit, Rat |
197 |
0.55 |
Binding ≤ 1μM
|
|
GBRA5_RAT |
P19969
|
GABA Receptor Alpha-5 Subunit, Rat |
197 |
0.55 |
Binding ≤ 1μM
|
|
GBRA6_RAT |
P30191
|
GABA Receptor Alpha-6 Subunit, Rat |
197 |
0.55 |
Binding ≤ 1μM
|
|
GBRB1_RAT |
P15431
|
GABA Receptor Beta-1 Subunit, Rat |
197 |
0.55 |
Binding ≤ 1μM
|
|
GBRB2_RAT |
P63138
|
GABA Receptor Beta-2 Subunit, Rat |
197 |
0.55 |
Binding ≤ 1μM
|
|
GBRB3_RAT |
P63079
|
GABA Receptor Beta-3 Subunit, Rat |
197 |
0.55 |
Binding ≤ 1μM
|
|
GBRD_RAT |
P18506
|
GABA Receptor Delta Subunit, Rat |
197 |
0.55 |
Binding ≤ 1μM
|
|
GBRE_RAT |
Q9ES14
|
GABA Receptor Epsilon Subunit, Rat |
197 |
0.55 |
Binding ≤ 1μM
|
|
GBRG1_RAT |
P23574
|
GABA Receptor Gamma-1 Subunit, Rat |
197 |
0.55 |
Binding ≤ 1μM
|
|
GBRG2_RAT |
P18508
|
GABA Receptor Gamma-2 Subunit, Rat |
197 |
0.55 |
Binding ≤ 1μM
|
|
GBRP_RAT |
O09028
|
GABA Receptor Pi Subunit, Rat |
197 |
0.55 |
Binding ≤ 1μM
|
|
GBRR1_RAT |
P50572
|
GABA Receptor Rho-1 Subunit, Rat |
197 |
0.55 |
Binding ≤ 1μM
|
|
GBRR2_RAT |
P47742
|
GABA Receptor Rho-2 Subunit, Rat |
197 |
0.55 |
Binding ≤ 1μM
|
|
Q91ZM7_RAT |
Q91ZM7
|
GABA Receptor Theta Subunit, Rat |
197 |
0.55 |
Binding ≤ 1μM
|
|
GBRA1_MOUSE |
P62812
|
GABA Receptor Alpha-1 Subunit, Mouse |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRA2_RAT |
P23576
|
GABA Receptor Alpha-2 Subunit, Rat |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRA3_RAT |
P20236
|
GABA Receptor Alpha-3 Subunit, Rat |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRA4_RAT |
P28471
|
GABA Receptor Alpha-4 Subunit, Rat |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRA5_RAT |
P19969
|
GABA Receptor Alpha-5 Subunit, Rat |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRA6_RAT |
P30191
|
GABA Receptor Alpha-6 Subunit, Rat |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRB1_RAT |
P15431
|
GABA Receptor Beta-1 Subunit, Rat |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRB2_RAT |
P63138
|
GABA Receptor Beta-2 Subunit, Rat |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRB3_RAT |
P63079
|
GABA Receptor Beta-3 Subunit, Rat |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRD_RAT |
P18506
|
GABA Receptor Delta Subunit, Rat |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRE_RAT |
Q9ES14
|
GABA Receptor Epsilon Subunit, Rat |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRG1_RAT |
P23574
|
GABA Receptor Gamma-1 Subunit, Rat |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRG2_RAT |
P18508
|
GABA Receptor Gamma-2 Subunit, Rat |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRP_RAT |
O09028
|
GABA Receptor Pi Subunit, Rat |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRR1_RAT |
P50572
|
GABA Receptor Rho-1 Subunit, Rat |
197 |
0.55 |
Binding ≤ 10μM
|
|
GBRR2_RAT |
P47742
|
GABA Receptor Rho-2 Subunit, Rat |
197 |
0.55 |
Binding ≤ 10μM
|
|
Q91ZM7_RAT |
Q91ZM7
|
GABA Receptor Theta Subunit, Rat |
197 |
0.55 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.02 |
3.27 |
-10.64 |
2 |
4 |
0 |
58 |
225.251 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.02 |
3.76 |
-38.44 |
3 |
4 |
1 |
59 |
226.259 |
1 |
↓
|
|
|
|
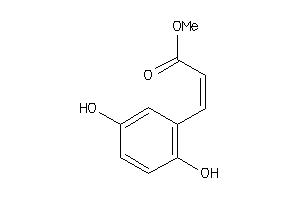
Analogs
-
13719806
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.78 |
0.75 |
-8.62 |
2 |
4 |
0 |
67 |
194.186 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.48 |
3.88 |
-10.98 |
3 |
5 |
0 |
93 |
308.337 |
4 |
↓
|
|
|
|
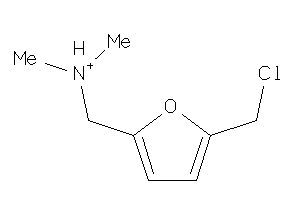
Analogs
-
39348609
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-5-E |
Muscarinic Acetylcholine Receptor M1 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
7943 |
0.65 |
Binding ≤ 10μM
|
|
ACM2-2-E |
Muscarinic Acetylcholine Receptor M2 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
955 |
0.77 |
Binding ≤ 10μM
|
|
ACM3-5-E |
Muscarinic Acetylcholine Receptor M3 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
6166 |
0.66 |
Binding ≤ 10μM
|
|
ACM4-4-E |
Muscarinic Acetylcholine Receptor M4 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
6310 |
0.66 |
Binding ≤ 10μM
|
|
ACM5-4-E |
Muscarinic Acetylcholine Receptor M5 (cluster #4 Of 4), Eukaryotic |
Eukaryotes |
6918 |
0.66 |
Binding ≤ 10μM
|
|
Z104303-7-O |
Muscarinic Acetylcholine Receptor (cluster #7 Of 7), Other |
Other |
6 |
1.05 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.56 |
7.34 |
-31.18 |
0 |
2 |
1 |
13 |
154.233 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A3-1-E |
Dopamine Transporter (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5340 |
0.32 |
Binding ≤ 10μM
|
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
44 |
0.45 |
Binding ≤ 10μM
|
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
5012 |
0.32 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.63 |
8.01 |
-5.93 |
0 |
4 |
0 |
39 |
317.429 |
6 |
↓
|
|
|
|
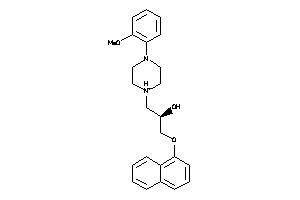
Analogs
-
597759
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.14 |
7.65 |
-9.6 |
1 |
5 |
0 |
45 |
392.499 |
7 |
↓
|
|
|
|
Analogs
-
597759
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.14 |
7.66 |
-9.68 |
1 |
5 |
0 |
45 |
392.499 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
700 |
0.36 |
Binding ≤ 10μM |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2500 |
0.33 |
Functional ≤ 10μM
|
|
NMDE2-1-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
660 |
0.36 |
Functional ≤ 10μM
|
|
NMDZ1-2-E |
Glutamate (NMDA) Receptor Subunit Zeta 1 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
660 |
0.36 |
Functional ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
7943 |
0.30 |
Functional ≤ 10μM
|
|
Z80951-1-O |
NIH3T3 (Fibroblasts) (cluster #1 Of 4), Other |
Other |
2500 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.85 |
4.57 |
-11.8 |
3 |
5 |
0 |
93 |
322.364 |
6 |
↓
|
|
|
|
Analogs
-
12352660
-
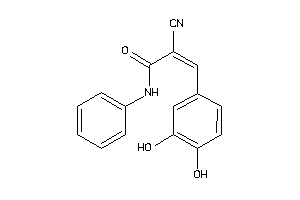
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
700 |
0.41 |
Binding ≤ 10μM |
|
PGFRA-1-E |
Platelet-derived Growth Factor Receptor Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.33 |
Binding ≤ 10μM
|
|
PGFRB-1-E |
Platelet-derived Growth Factor Receptor Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.33 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.22 |
2.9 |
-11.76 |
3 |
5 |
0 |
93 |
280.283 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LOX5-1-E |
Arachidonate 5-lipoxygenase (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
90 |
0.45 |
Binding ≤ 10μM
|
|
LX12L-1-E |
Arachidonate 12-lipoxygenase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
13 |
0.50 |
Binding ≤ 10μM
|
|
LX15B-1-E |
Arachidonate 15-lipoxygenase, Type II (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
500 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.98 |
5.48 |
-12.25 |
2 |
5 |
0 |
91 |
315.35 |
6 |
↓
|
|
|
|
Analogs
-
1530947
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH1-3-E |
Cyclooxygenase-1 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
370 |
0.47 |
Binding ≤ 10μM
|
|
PGH2-3-E |
Cyclooxygenase-2 (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
500 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.34 |
2.46 |
-13.39 |
1 |
6 |
0 |
85 |
258.233 |
1 |
↓
|
|
|
|
Analogs
-
1530948
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH1-3-E |
Cyclooxygenase-1 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
370 |
0.47 |
Binding ≤ 10μM
|
|
PGH2-3-E |
Cyclooxygenase-2 (cluster #3 Of 8), Eukaryotic |
Eukaryotes |
500 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.34 |
2.64 |
-15.5 |
1 |
6 |
0 |
85 |
258.233 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TSPO-1-E |
Peripheral-type Benzodiazepine Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.36 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.92 |
17.85 |
-8.98 |
1 |
3 |
0 |
36 |
487.515 |
13 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
2146111
-
-
4214170
-
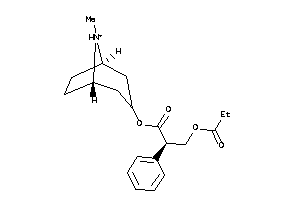
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACM1-4-E |
Muscarinic Acetylcholine Receptor M1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
3 |
0.50 |
Binding ≤ 10μM
|
|
ACM2-3-E |
Muscarinic Acetylcholine Receptor M2 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
2 |
0.51 |
Binding ≤ 10μM
|
|
ACM3-2-E |
Muscarinic Acetylcholine Receptor M3 (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
2 |
0.51 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.47 |
9.34 |
-34.51 |
1 |
4 |
1 |
47 |
332.464 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
4200 |
0.47 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
4200 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.87 |
6.53 |
-3.36 |
1 |
1 |
0 |
12 |
209.292 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1-1-E |
Sigma Opioid Receptor (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
4200 |
0.47 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SGMR1_RAT |
Q9R0C9
|
Sigma Opioid Receptor, Rat |
4200 |
0.47 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.87 |
6.49 |
-2.76 |
1 |
1 |
0 |
12 |
209.292 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT3A-1-E |
Serotonin 3a (5-HT3a) Receptor (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
950 |
0.34 |
Binding ≤ 10μM
|
|
5HT3B-1-E |
Serotonin 3b (5-HT3b) Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
950 |
0.34 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.78 |
7.78 |
-10.84 |
0 |
5 |
0 |
38 |
353.447 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTR1A_CHICK |
P49285
|
Melatonin Receptor 1A, Chick |
0.13 |
0.77 |
Binding ≤ 1μM
|
|
MTR1A_HUMAN |
P48039
|
Melatonin Receptor 1A, Human |
0.1 |
0.78 |
Binding ≤ 1μM
|
|
MTR1B_CHICK |
P51050
|
Melatonin Receptor 1B, Chick |
0.13 |
0.77 |
Binding ≤ 1μM
|
|
MTR1B_HUMAN |
P49286
|
Melatonin Receptor 1B, Human |
0.1 |
0.78 |
Binding ≤ 1μM
|
|
MTR1C_CHICK |
P49288
|
Melatonin Receptor 1C, Chick |
0.13 |
0.77 |
Binding ≤ 1μM
|
|
NQO2_HUMAN |
P16083
|
Quinone Reductase 2, Human |
6.5 |
0.64 |
Binding ≤ 1μM
|
|
MTR1A_CHICK |
P49285
|
Melatonin Receptor 1A, Chick |
0.13 |
0.77 |
Binding ≤ 10μM
|
|
MTR1A_HUMAN |
P48039
|
Melatonin Receptor 1A, Human |
0.1 |
0.78 |
Binding ≤ 10μM
|
|
MTR1B_CHICK |
P51050
|
Melatonin Receptor 1B, Chick |
0.13 |
0.77 |
Binding ≤ 10μM
|
|
MTR1B_HUMAN |
P49286
|
Melatonin Receptor 1B, Human |
0.1 |
0.78 |
Binding ≤ 10μM
|
|
MTR1C_CHICK |
P49288
|
Melatonin Receptor 1C, Chick |
0.13 |
0.77 |
Binding ≤ 10μM
|
|
NQO2_HUMAN |
P16083
|
Quinone Reductase 2, Human |
1100 |
0.46 |
Binding ≤ 10μM
|
|
MTR1A_HUMAN |
P48039
|
Melatonin Receptor 1A, Human |
0.1 |
0.78 |
Functional ≤ 10μM
|
|
MTR1B_HUMAN |
P49286
|
Melatonin Receptor 1B, Human |
0.1 |
0.78 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.65 |
4.5 |
-11.69 |
2 |
4 |
0 |
54 |
358.179 |
4 |
↓
|
|
|
|
Analogs
-
58590836
-
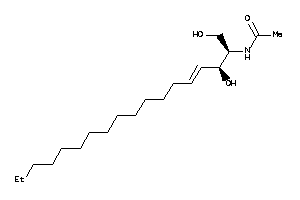
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.42 |
-3.21 |
-7.71 |
3 |
4 |
0 |
69 |
341.536 |
16 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7943 |
0.27 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
7943.28235 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.12 |
10.38 |
-9.37 |
0 |
4 |
0 |
34 |
350.462 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.13 |
6.89 |
-13.69 |
3 |
7 |
0 |
87 |
375.432 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.13 |
7.26 |
-29.57 |
4 |
7 |
1 |
88 |
376.44 |
6 |
↓
|
|