|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.44 |
-3.82 |
-11.37 |
4 |
7 |
0 |
99 |
303.366 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.44 |
-3.62 |
-38.89 |
5 |
7 |
1 |
100 |
304.374 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.44 |
-3.63 |
-39.25 |
5 |
7 |
1 |
100 |
304.374 |
5 |
↓
|
|
|
|
Analogs
-
2038619
-
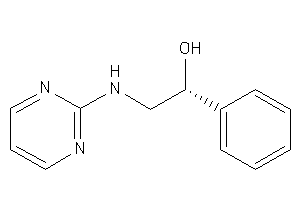
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.21 |
-3.61 |
-7.62 |
2 |
4 |
0 |
58 |
215.256 |
4 |
↓
|
|
|
|
Analogs
-
13984256
-
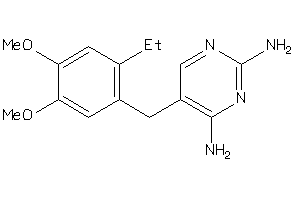
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RX2-1-E |
P2X Purinoceptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.35 |
Functional ≤ 10μM
|
|
P2RX3-1-E |
P2X Purinoceptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.39 |
4.67 |
-10.73 |
4 |
6 |
0 |
96 |
274.324 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.39 |
5.17 |
-38.05 |
5 |
6 |
1 |
98 |
275.332 |
4 |
↓
|
|
|
|
Analogs
-
3959
-
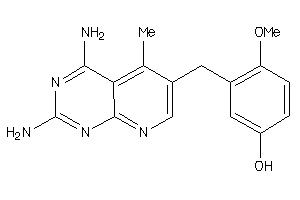
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
38 |
0.43 |
Binding ≤ 10μM
|
|
O30463-1-B |
Dihydrofolate Reductase (cluster #1 Of 1), Bacterial |
Bacteria |
1 |
0.53 |
Binding ≤ 10μM
|
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
43 |
0.43 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.49 |
Binding ≤ 10μM
|
|
DYR-1-F |
Dihydrofolate Reductase (cluster #1 Of 1), Fungal |
Fungi |
2 |
0.51 |
Binding ≤ 10μM
|
|
Z50339-1-O |
Pneumocystis Carinii (cluster #1 Of 2), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50472-1-O |
Toxoplasma Gondii (cluster #1 Of 4), Other |
Other |
20 |
0.45 |
Functional ≤ 10μM
|
|
Z80152-1-O |
HCT-8 (Ileocecal Adenocarcinoma) (cluster #1 Of 2), Other |
Other |
68 |
0.42 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
21 |
0.45 |
Functional ≤ 10μM
|
|
Z80563-1-O |
U-373 MG ( Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
55 |
0.42 |
Functional ≤ 10μM
|
|
Z80565-1-O |
U-87 MG (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
48 |
0.43 |
Functional ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 7), Other |
Other |
30 |
0.44 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
24 |
0.44 |
Functional ≤ 10μM
|
|
Z80815-1-O |
D54 Cell Line (cluster #1 Of 1), Other |
Other |
230 |
0.39 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
260 |
0.38 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS_TOXGO |
Q07422
|
Dihydrofolate Reductase, Toxgo |
10 |
0.47 |
Binding ≤ 1μM
|
|
DYR_CANAX |
P22906
|
Dihydrofolate Reductase, Canax |
1.9 |
0.51 |
Binding ≤ 1μM
|
|
DYR_PNECA |
P16184
|
Dihydrofolate Reductase, Pneca |
0.143 |
0.57 |
Binding ≤ 1μM
|
|
DYR_ECOLI |
P0ABQ4
|
Dihydrofolate Reductase, Ecoli |
11 |
0.46 |
Binding ≤ 1μM
|
|
DYR_YEAST |
P07807
|
Dihydrofolate Reductase, Yeast |
31 |
0.44 |
Binding ≤ 1μM
|
|
DYR_LACCA |
P00381
|
Dihydrofolate Reductase, Lacca |
1.5 |
0.51 |
Binding ≤ 1μM
|
|
DYR_HUMAN |
P00374
|
Dihydrofolate Reductase, Human |
0.1 |
0.58 |
Binding ≤ 1μM
|
|
O30463_MYCAV |
O30463
|
Dihydrofolate Reductase, Mycav |
0.61 |
0.54 |
Binding ≤ 1μM
|
|
DYR_RAT |
Q920D2
|
Dihydrofolate Reductase, Rat |
1 |
0.53 |
Binding ≤ 1μM
|
|
DYR_PNECA |
P16184
|
Dihydrofolate Reductase, Pneca |
0.143 |
0.57 |
Binding ≤ 10μM
|
|
DYR_ECOLI |
P0ABQ4
|
Dihydrofolate Reductase, Ecoli |
11 |
0.46 |
Binding ≤ 10μM
|
|
DYR_YEAST |
P07807
|
Dihydrofolate Reductase, Yeast |
31 |
0.44 |
Binding ≤ 10μM
|
|
DYR_LACCA |
P00381
|
Dihydrofolate Reductase, Lacca |
1.5 |
0.51 |
Binding ≤ 10μM
|
|
DYR_HUMAN |
P00374
|
Dihydrofolate Reductase, Human |
0.1 |
0.58 |
Binding ≤ 10μM
|
|
O30463_MYCAV |
O30463
|
Dihydrofolate Reductase, Mycav |
0.61 |
0.54 |
Binding ≤ 10μM
|
|
DYR_RAT |
Q920D2
|
Dihydrofolate Reductase, Rat |
1 |
0.53 |
Binding ≤ 10μM
|
|
DRTS_TOXGO |
Q07422
|
Dihydrofolate Reductase, Toxgo |
10 |
0.47 |
Binding ≤ 10μM
|
|
DYR_CANAX |
P22906
|
Dihydrofolate Reductase, Canax |
1.9 |
0.51 |
Binding ≤ 10μM
|
|
Z80682 |
Z80682
|
A549 (Lung Carcinoma Cells) |
24 |
0.44 |
Functional ≤ 10μM
|
|
Z80815 |
Z80815
|
D54 Cell Line |
230 |
0.39 |
Functional ≤ 10μM
|
|
Z80152 |
Z80152
|
HCT-8 (Ileocecal Adenocarcinoma) |
68 |
0.42 |
Functional ≤ 10μM
|
|
Z81024 |
Z81024
|
NCI-H460 (Non-small Cell Lung Carcinoma) |
260 |
0.38 |
Functional ≤ 10μM
|
|
Z80362 |
Z80362
|
P388 (Lymphoma Cells) |
21 |
0.45 |
Functional ≤ 10μM
|
|
Z50339 |
Z50339
|
Pneumocystis Carinii |
0.1 |
0.58 |
Functional ≤ 10μM
|
|
Z50472 |
Z50472
|
Toxoplasma Gondii |
0.1 |
0.58 |
Functional ≤ 10μM
|
|
Z80563 |
Z80563
|
U-373 MG ( Glioblastoma Cells) |
55 |
0.42 |
Functional ≤ 10μM
|
|
Z80565 |
Z80565
|
U-87 MG (Glioblastoma Cells) |
48 |
0.43 |
Functional ≤ 10μM
|
|
Z80583 |
Z80583
|
Vero (Kidney Cells) |
30 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
-3.63 |
-16.02 |
4 |
7 |
0 |
109 |
325.372 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.41 |
-3.41 |
-41.04 |
5 |
7 |
1 |
110 |
326.38 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.41 |
-3.4 |
-28.87 |
5 |
7 |
1 |
110 |
326.38 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.76 |
9.01 |
-13.11 |
0 |
8 |
0 |
73 |
467.039 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.76 |
11.23 |
-49.65 |
1 |
8 |
1 |
74 |
468.047 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.65 |
11.07 |
-12.41 |
0 |
7 |
0 |
70 |
452.024 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.65 |
11.34 |
-13.12 |
0 |
7 |
0 |
70 |
452.024 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.78 |
9.34 |
-13.83 |
0 |
9 |
0 |
88 |
496.033 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.78 |
9.61 |
-14.47 |
0 |
9 |
0 |
88 |
496.033 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.84 |
9.07 |
-19.31 |
0 |
10 |
0 |
99 |
511.048 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.84 |
9.03 |
-19.09 |
0 |
10 |
0 |
99 |
511.048 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.89 |
8.14 |
-21.68 |
1 |
8 |
0 |
91 |
475.018 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.89 |
8.52 |
-47.93 |
2 |
8 |
1 |
93 |
476.026 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.89 |
8.12 |
-21.29 |
1 |
8 |
0 |
91 |
475.018 |
8 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.89 |
8.5 |
-47.78 |
2 |
8 |
1 |
93 |
476.026 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.62 |
-5.75 |
-15.02 |
1 |
7 |
0 |
78 |
409.943 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.62 |
-5.75 |
-14.84 |
1 |
7 |
0 |
78 |
409.943 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.89 |
12.59 |
-13.17 |
0 |
7 |
0 |
70 |
486.041 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.89 |
12.86 |
-13.91 |
0 |
7 |
0 |
70 |
486.041 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.54 |
8.93 |
-15.57 |
1 |
9 |
0 |
97 |
504.012 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.07 |
10.25 |
-12.68 |
1 |
7 |
0 |
78 |
463.966 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.11 |
9.08 |
-18.65 |
1 |
7 |
0 |
78 |
463.966 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.11 |
9.07 |
-18.6 |
1 |
7 |
0 |
78 |
463.966 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.49 |
11.55 |
-15.63 |
0 |
8 |
0 |
73 |
529.11 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.49 |
11.53 |
-15.47 |
0 |
8 |
0 |
73 |
529.11 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.36 |
8.5 |
-14.29 |
1 |
8 |
0 |
91 |
460.991 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.36 |
8.96 |
-47.1 |
2 |
8 |
1 |
93 |
461.999 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.36 |
8.78 |
-15.05 |
1 |
8 |
0 |
91 |
460.991 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.36 |
9.23 |
-47.89 |
2 |
8 |
1 |
93 |
461.999 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.75 |
8.32 |
-20.04 |
0 |
9 |
0 |
90 |
481.022 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.75 |
8.29 |
-22.72 |
0 |
9 |
0 |
90 |
481.022 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.37 |
8.06 |
-17.68 |
1 |
8 |
0 |
91 |
448.98 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.37 |
8.52 |
-50.14 |
2 |
8 |
1 |
93 |
449.988 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.37 |
7.84 |
-16.96 |
1 |
8 |
0 |
91 |
448.98 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.37 |
8.3 |
-49.56 |
2 |
8 |
1 |
93 |
449.988 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
9.17 |
-16.72 |
1 |
7 |
0 |
78 |
477.993 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
9.16 |
-16.65 |
1 |
7 |
0 |
78 |
477.993 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
7.32 |
-17.61 |
1 |
8 |
0 |
91 |
460.991 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.41 |
7.75 |
-51.7 |
2 |
8 |
1 |
93 |
461.999 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
7.34 |
-17.66 |
1 |
8 |
0 |
91 |
460.991 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.41 |
7.77 |
-51.78 |
2 |
8 |
1 |
93 |
461.999 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.48 |
8.43 |
-14.46 |
1 |
8 |
0 |
91 |
460.991 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.48 |
8.83 |
-44.34 |
2 |
8 |
1 |
93 |
461.999 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.48 |
8.7 |
-15.16 |
1 |
8 |
0 |
91 |
460.991 |
7 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.48 |
9.11 |
-45.13 |
2 |
8 |
1 |
93 |
461.999 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.89 |
10.03 |
-14.26 |
1 |
7 |
0 |
78 |
480.009 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.89 |
10.3 |
-13.11 |
1 |
7 |
0 |
78 |
480.009 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.44 |
9.32 |
-14.6 |
1 |
7 |
0 |
78 |
465.982 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.44 |
9.59 |
-13.44 |
1 |
7 |
0 |
78 |
465.982 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.67 |
11.2 |
-14.5 |
1 |
7 |
0 |
78 |
506.047 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.67 |
11.47 |
-13.28 |
1 |
7 |
0 |
78 |
506.047 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.16 |
10.38 |
-14.66 |
1 |
7 |
0 |
78 |
514.026 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.16 |
10.35 |
-19.09 |
1 |
7 |
0 |
78 |
514.026 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.92 |
11.72 |
-14.35 |
0 |
7 |
0 |
70 |
492.02 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.92 |
11.99 |
-13.19 |
0 |
7 |
0 |
70 |
492.02 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.41 |
10.94 |
-14.95 |
0 |
7 |
0 |
70 |
477.993 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.41 |
11.21 |
-13.79 |
0 |
7 |
0 |
70 |
477.993 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.85 |
8.16 |
-18.92 |
0 |
8 |
0 |
79 |
493.992 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.85 |
8.2 |
-19.04 |
0 |
8 |
0 |
79 |
493.992 |
5 |
↓
|
|