|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.24 |
0.25 |
-92.56 |
6 |
10 |
-1 |
188 |
443.432 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.69 |
1.2 |
-176.8 |
5 |
10 |
-2 |
191 |
442.424 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.69 |
-2.17 |
-117.19 |
5 |
10 |
-2 |
187 |
442.424 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.24 |
0.59 |
-95.46 |
6 |
10 |
-1 |
188 |
443.432 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.69 |
-1.58 |
-146.85 |
5 |
10 |
-2 |
187 |
442.424 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.24 |
-0.62 |
-247.57 |
4 |
10 |
-3 |
190 |
441.416 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.69 |
0.03 |
-52.94 |
7 |
10 |
0 |
186 |
444.44 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.69 |
-0.61 |
-112.68 |
5 |
10 |
-2 |
187 |
442.424 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.69 |
2.1 |
-172.87 |
5 |
10 |
-2 |
191 |
442.424 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.69 |
-0.49 |
-62.02 |
7 |
10 |
0 |
186 |
444.44 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.69 |
-1.06 |
-114.48 |
5 |
10 |
-2 |
187 |
442.424 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.69 |
1.6 |
-188.44 |
5 |
10 |
-2 |
191 |
442.424 |
2 |
↓
|
|
|
|
Analogs
-
9339994
-
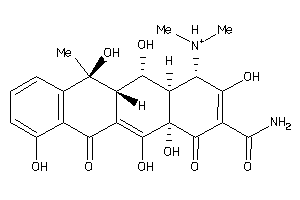
-
11592622
-
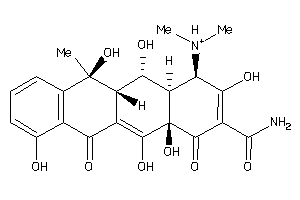
-
11592623
-
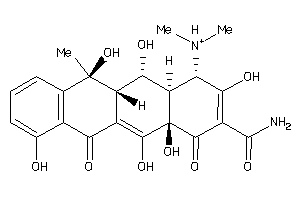
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4S,5S,6S,12aS)-4-(dimethylamino)-3,5,6,10,12,12a-hexahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide hydrochloride
(4S,5S,6S,12aS)-4-(dimethylamino…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.61 |
-2.42 |
-52.69 |
8 |
11 |
0 |
206 |
460.439 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.61 |
-3.75 |
-118.05 |
6 |
11 |
-2 |
208 |
458.423 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.34 |
-0.72 |
-152.03 |
7 |
11 |
-1 |
209 |
459.431 |
1 |
↓
|
|
|
|
Analogs
-
4879584
-
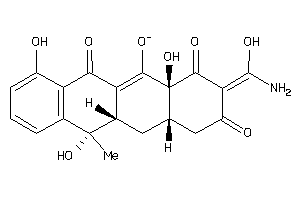
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.16 |
-2.23 |
-94.38 |
7 |
11 |
-1 |
209 |
459.431 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
-1.89 |
-4.35 |
-71.51 |
7 |
11 |
-1 |
205 |
459.431 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.34 |
-4.44 |
-70.79 |
7 |
11 |
-1 |
205 |
459.431 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4S,5S,6S,12aS)-4-(dimethylamino)-3,5,6,10,12,12a-hexahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide hydrochloride
(4S,5S,6S,12aS)-4-(dimethylamino…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.16 |
-0.07 |
-66.25 |
7 |
11 |
0 |
209 |
459.431 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
-1.61 |
-1.5 |
-23.92 |
7 |
11 |
0 |
205 |
459.431 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.34 |
-1.59 |
-73.05 |
7 |
11 |
-1 |
205 |
459.431 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(4S,5S,6S,12aS)-4-(dimethylamino)-3,5,6,10,12,12a-hexahydroxy-6-methyl-1,11-dioxo-1,4,4a,5,5a,6,11,12a-octahydrotetracene-2-carboxamide hydrochloride
(4S,5S,6S,12aS)-4-(dimethylamino…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.16 |
-0.1 |
-98.19 |
7 |
11 |
-1 |
209 |
459.431 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
-1.61 |
-2.81 |
-62.86 |
7 |
11 |
-1 |
205 |
459.431 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.61 |
-1.81 |
-117.35 |
6 |
11 |
-2 |
208 |
458.423 |
2 |
↓
|
|
|
|
Analogs
-
36372561
-
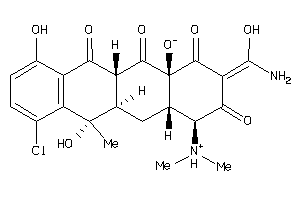
-
36372564
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.26 |
-3.99 |
-50.32 |
6 |
10 |
-1 |
185 |
477.877 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
-0.06 |
-0.42 |
-53.8 |
7 |
10 |
0 |
186 |
478.885 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.26 |
-3 |
-106.41 |
5 |
10 |
-2 |
188 |
476.869 |
2 |
↓
|
|
|
|
|
|
|
Analogs
-
38734506
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.39 |
0.69 |
-111.73 |
6 |
10 |
-1 |
188 |
477.877 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.06 |
-0.74 |
-106.71 |
5 |
10 |
-2 |
187 |
476.869 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.06 |
0.12 |
-234.75 |
4 |
10 |
-3 |
190 |
475.861 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.87 |
-1.65 |
-26.73 |
6 |
10 |
0 |
184 |
443.432 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.43 |
1.59 |
-100.24 |
6 |
10 |
0 |
188 |
443.432 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.43 |
-0.51 |
-71.63 |
5 |
10 |
0 |
187 |
442.424 |
2 |
↓
|
|
|
|
Analogs
-
16248873
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.43 |
0.98 |
-89.55 |
6 |
10 |
-1 |
188 |
443.432 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
-0.56 |
-4.34 |
-53.29 |
6 |
10 |
-1 |
185 |
443.432 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.61 |
-2.24 |
-69.99 |
6 |
10 |
-1 |
184 |
443.432 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.87 |
1.58 |
-25.31 |
6 |
10 |
0 |
184 |
443.432 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.43 |
1.69 |
-80.46 |
5 |
10 |
0 |
187 |
442.424 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.87 |
2.31 |
-51.6 |
7 |
10 |
0 |
186 |
444.44 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.36 |
2.01 |
-15.33 |
2 |
7 |
0 |
104 |
297.336 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
80 |
0.50 |
Binding ≤ 10μM
|
|
CHLE-1-E |
Butyrylcholinesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
280 |
0.46 |
Binding ≤ 10μM
|
|
Q9JKC1-1-E |
Butyrylcholinesterase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.57 |
Binding ≤ 10μM |
|
ACES-1-E |
Acetylcholinesterase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
128 |
0.48 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
6310 |
0.36 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES_RAT |
P37136
|
Acetylcholinesterase, Rat |
0.68 |
0.64 |
Binding ≤ 1μM
|
|
ACES_BOVIN |
P23795
|
Acetylcholinesterase, Bovin |
123 |
0.48 |
Binding ≤ 1μM
|
|
ACES_HUMAN |
P22303
|
Acetylcholinesterase, Human |
28 |
0.53 |
Binding ≤ 1μM
|
|
ACES_MOUSE |
P21836
|
Acetylcholinesterase, Mouse |
0.68 |
0.64 |
Binding ≤ 1μM
|
|
ACES_TORCA |
P04058
|
Acetylcholinesterase, Torca |
21.3 |
0.54 |
Binding ≤ 1μM
|
|
ACES_ELEEL |
O42275
|
Acetylcholinesterase, Eleel |
1000 |
0.42 |
Binding ≤ 1μM
|
|
CHLE_MOUSE |
Q03311
|
Butyrylcholinesterase, Mouse |
280 |
0.46 |
Binding ≤ 1μM
|
|
CHLE_HUMAN |
P06276
|
Butyrylcholinesterase, Human |
16 |
0.55 |
Binding ≤ 1μM
|
|
Q9JKC1_RAT |
Q9JKC1
|
Butyrylcholinesterase, Rat |
23 |
0.53 |
Binding ≤ 1μM
|
|
CHLE_HORSE |
P81908
|
Cholinesterase, Horse |
857 |
0.42 |
Binding ≤ 1μM
|
|
ACES_RAT |
P37136
|
Acetylcholinesterase, Rat |
0.68 |
0.64 |
Binding ≤ 10μM
|
|
ACES_BOVIN |
P23795
|
Acetylcholinesterase, Bovin |
123 |
0.48 |
Binding ≤ 10μM
|
|
ACES_HUMAN |
P22303
|
Acetylcholinesterase, Human |
28 |
0.53 |
Binding ≤ 10μM
|
|
ACES_MOUSE |
P21836
|
Acetylcholinesterase, Mouse |
0.68 |
0.64 |
Binding ≤ 10μM
|
|
ACES_TORCA |
P04058
|
Acetylcholinesterase, Torca |
21.3 |
0.54 |
Binding ≤ 10μM
|
|
ACES_ELEEL |
O42275
|
Acetylcholinesterase, Eleel |
1000 |
0.42 |
Binding ≤ 10μM
|
|
CHLE_MOUSE |
Q03311
|
Butyrylcholinesterase, Mouse |
280 |
0.46 |
Binding ≤ 10μM
|
|
CHLE_HUMAN |
P06276
|
Butyrylcholinesterase, Human |
16 |
0.55 |
Binding ≤ 10μM
|
|
Q9JKC1_RAT |
Q9JKC1
|
Butyrylcholinesterase, Rat |
23 |
0.53 |
Binding ≤ 10μM
|
|
CHLE_HORSE |
P81908
|
Cholinesterase, Horse |
1340 |
0.41 |
Binding ≤ 10μM
|
|
ACES_HUMAN |
P22303
|
Acetylcholinesterase, Human |
128 |
0.48 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
3981.07171 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.90 |
-0.07 |
-7.08 |
1 |
5 |
0 |
48 |
275.352 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.90 |
2.61 |
-39.01 |
2 |
5 |
1 |
50 |
276.36 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.90 |
4.54 |
-76.78 |
3 |
5 |
2 |
51 |
277.368 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.77 |
-3.12 |
-135.84 |
2 |
6 |
-2 |
113 |
168.041 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
-2.45 |
-1.03 |
-138.28 |
1 |
6 |
-2 |
110 |
168.041 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
-1.81 |
-1.29 |
-130.45 |
1 |
6 |
-2 |
110 |
168.041 |
4 |
↓
|
|
|
|
Analogs
-
35024421
-
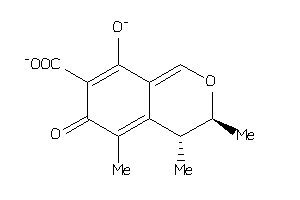
-
1164
-
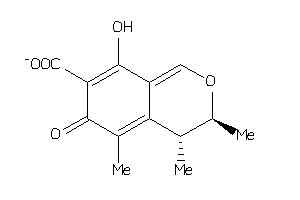
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.48 |
6.5 |
-159.38 |
0 |
5 |
-2 |
89 |
248.234 |
1 |
↓
|
|
Ref
Reference (pH 7)
|
1.48 |
6.49 |
-157.53 |
0 |
5 |
-2 |
89 |
248.234 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.48 |
4.5 |
-54.79 |
1 |
5 |
-1 |
87 |
249.242 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.24 |
6.4 |
-6.75 |
1 |
4 |
0 |
54 |
207.277 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.21 |
3.05 |
-10.35 |
1 |
5 |
0 |
71 |
258.362 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.21 |
4.48 |
-9.96 |
1 |
5 |
0 |
71 |
258.362 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.21 |
4.49 |
-9.89 |
1 |
5 |
0 |
71 |
258.362 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.21 |
3.02 |
-10.33 |
1 |
5 |
0 |
71 |
258.362 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
200 |
0.49 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80193 |
Z80193
|
L1210 (Lymphocytic Leukemia Cells) |
1000 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.31 |
-6.92 |
-15.02 |
6 |
9 |
0 |
150 |
267.245 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.31 |
-11.19 |
-40.6 |
7 |
9 |
1 |
152 |
268.253 |
2 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.94 |
-5.66 |
-15.21 |
4 |
7 |
0 |
124 |
222.193 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.74 |
4.12 |
-16.76 |
4 |
6 |
0 |
115 |
376.364 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.74 |
5.13 |
-66.02 |
3 |
6 |
-1 |
118 |
375.356 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.56 |
7.4 |
-123 |
1 |
6 |
-2 |
118 |
374.348 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.57 |
-1.13 |
-32.75 |
3 |
5 |
0 |
94 |
131.087 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.57 |
-2.77 |
-40.22 |
2 |
5 |
-1 |
99 |
130.079 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.57 |
-0.76 |
-100.08 |
1 |
5 |
-2 |
102 |
129.071 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.16 |
-4.21 |
-17.62 |
3 |
4 |
0 |
78 |
134.131 |
4 |
↓
|
|
|
|
Analogs
-
902219
-
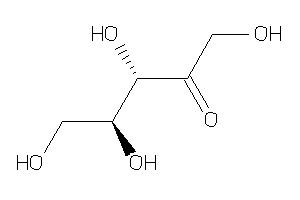
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.16 |
-4.21 |
-17.62 |
3 |
4 |
0 |
78 |
134.131 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Mid
Mid (pH 6-8)
|
1.53 |
2.84 |
-56.8 |
4 |
11 |
-1 |
189 |
475.437 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.34 |
-1.73 |
-54.55 |
3 |
6 |
-1 |
112 |
193.566 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.34 |
-2.62 |
-103.81 |
2 |
6 |
-2 |
111 |
192.558 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-3.34 |
-2.47 |
-104.8 |
2 |
6 |
-2 |
111 |
192.558 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.10 |
1.54 |
-36.69 |
2 |
3 |
0 |
66 |
101.105 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.10 |
1.63 |
-39.46 |
1 |
3 |
-1 |
64 |
100.097 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.10 |
1.63 |
-40.3 |
1 |
3 |
-1 |
64 |
100.097 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50459-1-O |
Leishmania Donovani (cluster #1 Of 8), Other |
Other |
80 |
0.34 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.70 |
2.16 |
-101.92 |
4 |
9 |
0 |
150 |
391.339 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.70 |
3.18 |
-104.89 |
3 |
9 |
-1 |
153 |
390.331 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.70 |
2.69 |
-127.35 |
2 |
9 |
-2 |
151 |
389.323 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.35 |
-0.29 |
-23.86 |
6 |
12 |
0 |
192 |
571.579 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.35 |
0.08 |
-23.93 |
6 |
12 |
0 |
192 |
571.579 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.35 |
0.03 |
-23.83 |
6 |
12 |
0 |
192 |
571.579 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.35 |
0.47 |
-23.54 |
6 |
12 |
0 |
192 |
571.579 |
5 |
↓
|
|
|
|
Analogs
-
4403394
-
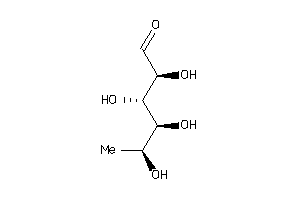
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.64 |
-6.46 |
-8.59 |
4 |
5 |
0 |
98 |
164.157 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.49 |
-5.34 |
-54.76 |
3 |
5 |
-1 |
101 |
163.149 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.49 |
-5.37 |
-54.67 |
3 |
5 |
-1 |
101 |
163.149 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.77 |
11.08 |
-15.89 |
0 |
6 |
0 |
85 |
341.363 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.91 |
17.85 |
-67 |
3 |
9 |
-1 |
153 |
708.01 |
23 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50725-6-O |
Trypanosoma Brucei Rhodesiense (cluster #6 Of 7), Other |
Other |
3710 |
0.40 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50725 |
Z50725
|
Trypanosoma Brucei Rhodesiense |
3710 |
0.40 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ECE1-1-E |
Endothelin-converting Enzyme 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
800 |
0.23 |
Binding ≤ 10μM
|
|
NEP-2-E |
Neprilysin (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.31 |
Binding ≤ 10μM
|
|
THER-1-B |
Thermolysin (cluster #1 Of 3), Bacterial |
Bacteria |
33 |
0.28 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.56 |
-1.57 |
-111.49 |
6 |
13 |
-2 |
220 |
541.494 |
11 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-1.56 |
-0.31 |
-60.51 |
7 |
13 |
-1 |
221 |
542.502 |
11 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.69 |
2.98 |
-9.62 |
1 |
5 |
0 |
68 |
275.333 |
4 |
↓
|
|