|
|
Analogs
-
3959
-
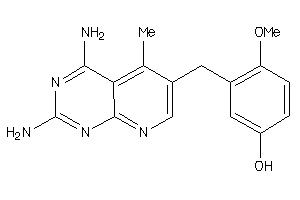
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
38 |
0.43 |
Binding ≤ 10μM
|
|
O30463-1-B |
Dihydrofolate Reductase (cluster #1 Of 1), Bacterial |
Bacteria |
1 |
0.53 |
Binding ≤ 10μM
|
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
43 |
0.43 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
4 |
0.49 |
Binding ≤ 10μM
|
|
DYR-1-F |
Dihydrofolate Reductase (cluster #1 Of 1), Fungal |
Fungi |
2 |
0.51 |
Binding ≤ 10μM
|
|
Z50339-1-O |
Pneumocystis Carinii (cluster #1 Of 2), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50472-1-O |
Toxoplasma Gondii (cluster #1 Of 4), Other |
Other |
20 |
0.45 |
Functional ≤ 10μM
|
|
Z80152-1-O |
HCT-8 (Ileocecal Adenocarcinoma) (cluster #1 Of 2), Other |
Other |
68 |
0.42 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
21 |
0.45 |
Functional ≤ 10μM
|
|
Z80563-1-O |
U-373 MG ( Glioblastoma Cells) (cluster #1 Of 1), Other |
Other |
55 |
0.42 |
Functional ≤ 10μM
|
|
Z80565-1-O |
U-87 MG (Glioblastoma Cells) (cluster #1 Of 2), Other |
Other |
48 |
0.43 |
Functional ≤ 10μM
|
|
Z80583-1-O |
Vero (Kidney Cells) (cluster #1 Of 7), Other |
Other |
30 |
0.44 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
24 |
0.44 |
Functional ≤ 10μM
|
|
Z80815-1-O |
D54 Cell Line (cluster #1 Of 1), Other |
Other |
230 |
0.39 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
260 |
0.38 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR_YEAST |
P07807
|
Dihydrofolate Reductase, Yeast |
31 |
0.44 |
Binding ≤ 1μM
|
|
DYR_LACCA |
P00381
|
Dihydrofolate Reductase, Lacca |
1.5 |
0.51 |
Binding ≤ 1μM
|
|
DYR_HUMAN |
P00374
|
Dihydrofolate Reductase, Human |
0.1 |
0.58 |
Binding ≤ 1μM
|
|
O30463_MYCAV |
O30463
|
Dihydrofolate Reductase, Mycav |
0.61 |
0.54 |
Binding ≤ 1μM
|
|
DYR_RAT |
Q920D2
|
Dihydrofolate Reductase, Rat |
1 |
0.53 |
Binding ≤ 1μM
|
|
DRTS_TOXGO |
Q07422
|
Dihydrofolate Reductase, Toxgo |
10 |
0.47 |
Binding ≤ 1μM
|
|
DYR_CANAX |
P22906
|
Dihydrofolate Reductase, Canax |
1.9 |
0.51 |
Binding ≤ 1μM
|
|
DYR_PNECA |
P16184
|
Dihydrofolate Reductase, Pneca |
0.143 |
0.57 |
Binding ≤ 1μM
|
|
DYR_ECOLI |
P0ABQ4
|
Dihydrofolate Reductase, Ecoli |
11 |
0.46 |
Binding ≤ 1μM
|
|
DYR_HUMAN |
P00374
|
Dihydrofolate Reductase, Human |
0.1 |
0.58 |
Binding ≤ 10μM
|
|
O30463_MYCAV |
O30463
|
Dihydrofolate Reductase, Mycav |
0.61 |
0.54 |
Binding ≤ 10μM
|
|
DYR_RAT |
Q920D2
|
Dihydrofolate Reductase, Rat |
1 |
0.53 |
Binding ≤ 10μM
|
|
DRTS_TOXGO |
Q07422
|
Dihydrofolate Reductase, Toxgo |
10 |
0.47 |
Binding ≤ 10μM
|
|
DYR_CANAX |
P22906
|
Dihydrofolate Reductase, Canax |
1.9 |
0.51 |
Binding ≤ 10μM
|
|
DYR_PNECA |
P16184
|
Dihydrofolate Reductase, Pneca |
0.143 |
0.57 |
Binding ≤ 10μM
|
|
DYR_ECOLI |
P0ABQ4
|
Dihydrofolate Reductase, Ecoli |
11 |
0.46 |
Binding ≤ 10μM
|
|
DYR_YEAST |
P07807
|
Dihydrofolate Reductase, Yeast |
31 |
0.44 |
Binding ≤ 10μM
|
|
DYR_LACCA |
P00381
|
Dihydrofolate Reductase, Lacca |
1.5 |
0.51 |
Binding ≤ 10μM
|
|
Z80682 |
Z80682
|
A549 (Lung Carcinoma Cells) |
24 |
0.44 |
Functional ≤ 10μM
|
|
Z80815 |
Z80815
|
D54 Cell Line |
230 |
0.39 |
Functional ≤ 10μM
|
|
Z80152 |
Z80152
|
HCT-8 (Ileocecal Adenocarcinoma) |
68 |
0.42 |
Functional ≤ 10μM
|
|
Z81024 |
Z81024
|
NCI-H460 (Non-small Cell Lung Carcinoma) |
260 |
0.38 |
Functional ≤ 10μM
|
|
Z80362 |
Z80362
|
P388 (Lymphoma Cells) |
21 |
0.45 |
Functional ≤ 10μM
|
|
Z50339 |
Z50339
|
Pneumocystis Carinii |
0.1 |
0.58 |
Functional ≤ 10μM
|
|
Z50472 |
Z50472
|
Toxoplasma Gondii |
0.1 |
0.58 |
Functional ≤ 10μM
|
|
Z80563 |
Z80563
|
U-373 MG ( Glioblastoma Cells) |
55 |
0.42 |
Functional ≤ 10μM
|
|
Z80565 |
Z80565
|
U-87 MG (Glioblastoma Cells) |
48 |
0.43 |
Functional ≤ 10μM
|
|
Z80583 |
Z80583
|
Vero (Kidney Cells) |
30 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
-3.63 |
-16.02 |
4 |
7 |
0 |
109 |
325.372 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.41 |
-3.41 |
-41.04 |
5 |
7 |
1 |
110 |
326.38 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.41 |
-3.4 |
-28.87 |
5 |
7 |
1 |
110 |
326.38 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADK-1-E |
Adenosine Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8 |
0.31 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
10.99 |
-18.52 |
3 |
9 |
0 |
127 |
561.44 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.50 |
4.48 |
-122.82 |
6 |
12 |
-2 |
212 |
437.416 |
9 |
↓
|
|
|
|
Analogs
-
17022483
-
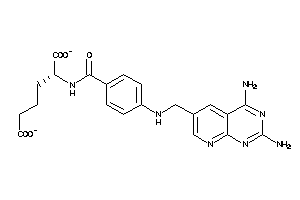
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.45 |
5.24 |
-122.47 |
6 |
12 |
-2 |
212 |
451.443 |
10 |
↓
|
|
|
|
Analogs
-
39386676
-
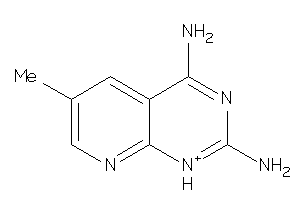
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.22 |
2.46 |
-12.58 |
4 |
6 |
0 |
115 |
186.178 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.22 |
2.78 |
-33.87 |
5 |
6 |
1 |
116 |
187.186 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.22 |
-2.95 |
-38 |
5 |
6 |
1 |
116 |
187.186 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.49 |
4.4 |
-30.93 |
0 |
4 |
-1 |
62 |
277.518 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.04 |
5.67 |
-10.02 |
1 |
4 |
0 |
59 |
278.526 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.00 |
3.15 |
-53.24 |
1 |
6 |
-1 |
87 |
238.23 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.54 |
5.23 |
-13.59 |
2 |
6 |
0 |
84 |
239.238 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
7.19 |
-43.88 |
0 |
6 |
-1 |
74 |
341.778 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTOR-2-E |
Serine/threonine-protein Kinase MTOR (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1 |
0.37 |
Binding ≤ 10μM |
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MTOR_HUMAN |
P42345
|
FK506 Binding Protein 12, Human |
0.8 |
0.37 |
Binding ≤ 1μM |
|
MTOR_HUMAN |
P42345
|
FK506 Binding Protein 12, Human |
0.8 |
0.37 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.39 |
9.2 |
-16.08 |
1 |
9 |
0 |
93 |
465.554 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.86 |
3.59 |
-56.87 |
2 |
8 |
-1 |
120 |
396.452 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.69 |
3.97 |
-48.76 |
1 |
7 |
-1 |
100 |
369.426 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.08 |
0.61 |
-117.61 |
0 |
7 |
-2 |
111 |
279.277 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.62 |
4.57 |
-42.81 |
1 |
7 |
-1 |
108 |
280.285 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.76 |
5.12 |
-55.36 |
0 |
8 |
-1 |
114 |
345.36 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.30 |
6.82 |
-17.75 |
1 |
8 |
0 |
111 |
346.368 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.89 |
2.72 |
-56.06 |
0 |
9 |
-1 |
127 |
346.348 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.44 |
4.41 |
-17.84 |
1 |
9 |
0 |
124 |
347.356 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.32 |
1.94 |
-54.45 |
1 |
8 |
-1 |
117 |
321.338 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.87 |
3.65 |
-18.37 |
2 |
8 |
0 |
114 |
322.346 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.13 |
3.94 |
-54.08 |
0 |
8 |
-1 |
108 |
335.365 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.67 |
5.64 |
-17.81 |
1 |
8 |
0 |
105 |
336.373 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.19 |
5.21 |
-56.21 |
0 |
8 |
-1 |
106 |
344.376 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.73 |
6.9 |
-17.97 |
1 |
8 |
0 |
103 |
345.384 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.94 |
9.6 |
-54.79 |
0 |
7 |
-1 |
105 |
424.502 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
5.83 |
-47.38 |
0 |
6 |
-1 |
82 |
359.475 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.29 |
7.59 |
-10.91 |
1 |
6 |
0 |
79 |
360.483 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
6.17 |
-47.6 |
0 |
6 |
-1 |
82 |
359.475 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.29 |
7.93 |
-10.77 |
1 |
6 |
0 |
79 |
360.483 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
6.17 |
-47.56 |
0 |
6 |
-1 |
82 |
359.475 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.29 |
7.93 |
-10.81 |
1 |
6 |
0 |
79 |
360.483 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.90 |
5.65 |
-47.74 |
1 |
6 |
-1 |
91 |
367.454 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.44 |
7.41 |
-12.57 |
2 |
6 |
0 |
88 |
368.462 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.90 |
5.65 |
-47.8 |
1 |
6 |
-1 |
91 |
367.454 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.44 |
7.41 |
-12.58 |
2 |
6 |
0 |
88 |
368.462 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.39 |
4.11 |
-47.67 |
1 |
6 |
-1 |
91 |
333.437 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.94 |
5.87 |
-12.08 |
2 |
6 |
0 |
88 |
334.445 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.39 |
3.81 |
-47.5 |
1 |
6 |
-1 |
91 |
333.437 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.94 |
5.56 |
-11.99 |
2 |
6 |
0 |
88 |
334.445 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.36 |
6.84 |
-60.3 |
0 |
9 |
-1 |
119 |
345.364 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.91 |
8.54 |
-21.68 |
1 |
9 |
0 |
116 |
346.372 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.54 |
0.31 |
-42.11 |
0 |
4 |
-1 |
62 |
180.574 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.40 |
7.26 |
-49.07 |
2 |
5 |
1 |
55 |
246.338 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.40 |
5.11 |
-12.16 |
1 |
5 |
0 |
54 |
245.33 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.19 |
7.45 |
-48.55 |
2 |
5 |
1 |
55 |
246.338 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.24 |
11.77 |
-48.77 |
2 |
6 |
1 |
58 |
377.516 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.75 |
7.85 |
-12.77 |
1 |
4 |
0 |
51 |
222.251 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.41 |
8.36 |
-11.95 |
1 |
4 |
0 |
51 |
256.696 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.41 |
11.13 |
-14.6 |
1 |
5 |
0 |
60 |
328.375 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.25 |
4.95 |
-14.86 |
2 |
5 |
0 |
71 |
238.25 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.27 |
4.97 |
-14.6 |
2 |
5 |
0 |
71 |
238.25 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.91 |
5.85 |
-12.53 |
2 |
5 |
0 |
71 |
252.277 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.14 |
5.79 |
-13.16 |
2 |
5 |
0 |
71 |
272.695 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.64 |
8.4 |
-64.13 |
1 |
6 |
-1 |
91 |
265.252 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.66 |
8.34 |
-57.4 |
1 |
6 |
-1 |
91 |
265.252 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.04 |
9.27 |
-64.67 |
1 |
6 |
-1 |
91 |
279.279 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.18 |
9.17 |
-53.58 |
1 |
6 |
-1 |
91 |
279.279 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.65 |
5.87 |
-14.37 |
2 |
5 |
0 |
71 |
252.277 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.52 |
7.22 |
-11.86 |
0 |
7 |
0 |
64 |
301.35 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.26 |
11.36 |
-127.97 |
7 |
12 |
-1 |
213 |
528.549 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.26 |
9.43 |
-80.78 |
8 |
12 |
0 |
211 |
529.557 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.16 |
5.06 |
-53.6 |
2 |
5 |
1 |
59 |
216.268 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.16 |
3.76 |
-10.43 |
1 |
5 |
0 |
54 |
215.26 |
1 |
↓
|
|