|
|
Analogs
-
22060777
-
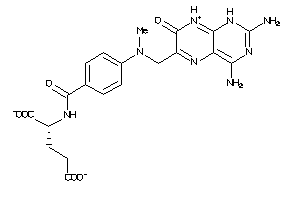
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.52 |
6 |
-128.06 |
7 |
14 |
-1 |
237 |
469.438 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.06 |
1.11 |
-178.14 |
5 |
14 |
-3 |
239 |
467.422 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.52 |
5.48 |
-119.99 |
6 |
14 |
-2 |
236 |
468.43 |
9 |
↓
|
|
|
|
Analogs
-
38494715
-
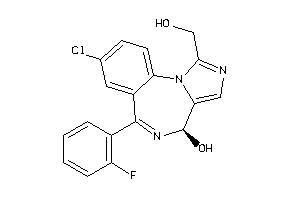
-
38494717
-
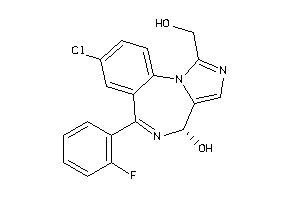
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.61 |
5.16 |
-32.8 |
2 |
4 |
1 |
52 |
342.781 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.61 |
4.7 |
-12.04 |
1 |
4 |
0 |
50 |
341.773 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Hi
High (pH 8-9.5)
|
2.33 |
4.39 |
-3.59 |
1 |
1 |
0 |
20 |
182.288 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.12 |
-2.18 |
-39.96 |
5 |
8 |
1 |
125 |
365.435 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.12 |
-2.09 |
-58.19 |
3 |
8 |
-1 |
127 |
363.419 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.12 |
-2.51 |
-13.27 |
4 |
8 |
0 |
124 |
364.427 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.30 |
4.49 |
-39.58 |
3 |
7 |
1 |
93 |
374.442 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.30 |
5.15 |
-54.43 |
1 |
7 |
-1 |
95 |
372.426 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.30 |
5.19 |
-31.29 |
2 |
7 |
0 |
96 |
373.434 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.30 |
4.44 |
-39.43 |
3 |
7 |
1 |
93 |
374.442 |
7 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.30 |
5.15 |
-57 |
1 |
7 |
-1 |
95 |
372.426 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.30 |
5.16 |
-31.5 |
2 |
7 |
0 |
96 |
373.434 |
7 |
↓
|
|
|
|
Analogs
-
509440
-
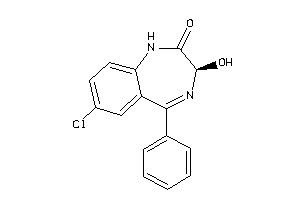
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.84 |
1.04 |
-8.23 |
2 |
4 |
0 |
62 |
286.718 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.02 |
-1.08 |
-50.39 |
1 |
4 |
-1 |
68 |
285.71 |
1 |
↓
|
|
|
|
Analogs
-
575
-
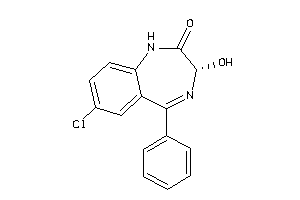
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.84 |
2.21 |
-11.02 |
2 |
4 |
0 |
62 |
286.718 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.02 |
0.08 |
-55.63 |
1 |
4 |
-1 |
68 |
285.71 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS-3-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
82 |
0.58 |
Binding ≤ 10μM
|
|
DYR-2-E |
Dihydrofolate Reductase (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
110 |
0.57 |
Binding ≤ 10μM
|
|
DRTS-1-E |
Dihydrofolate Reductase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5800 |
0.43 |
Functional ≤ 10μM
|
|
DYR-1-F |
Dihydrofolate Reductase (cluster #1 Of 1), Fungal |
Fungi |
450 |
0.52 |
Binding ≤ 10μM
|
|
Z50425-7-O |
Plasmodium Falciparum (cluster #7 Of 22), Other |
Other |
8 |
0.67 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.90 |
7.06 |
-25.38 |
5 |
5 |
1 |
82 |
252.729 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.16 |
6.71 |
-4.72 |
4 |
5 |
0 |
75 |
251.721 |
1 |
↓
|
|
|
|
|
|
|
Analogs
-
5923859
-
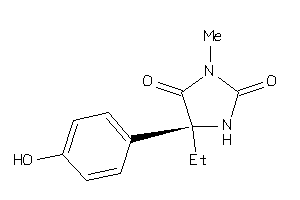
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.23 |
1.74 |
-8.92 |
2 |
5 |
0 |
70 |
234.255 |
2 |
↓
|
|
|
|
Analogs
-
2512178
-
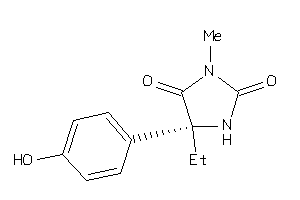
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.23 |
1.83 |
-9.39 |
2 |
5 |
0 |
70 |
234.255 |
2 |
↓
|
|
|
|
Analogs
-
546
-
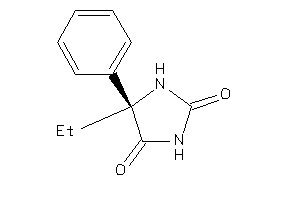
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.46 |
2.34 |
-7.44 |
2 |
4 |
0 |
58 |
204.229 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.65 |
-0.33 |
-44.3 |
1 |
4 |
-1 |
65 |
203.221 |
2 |
↓
|
|
|
|
Analogs
-
38438731
-
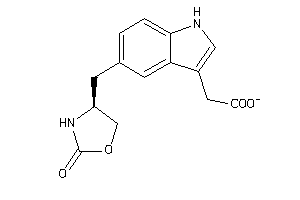
-
5996379
-
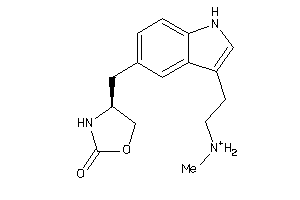
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.71 |
2 |
-49.44 |
5 |
5 |
1 |
82 |
260.317 |
4 |
↓
|
|
|
|
Analogs
-
30731123
-
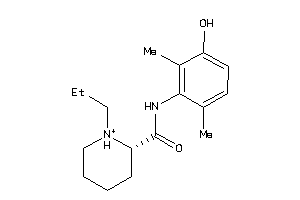
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.85 |
4.46 |
-35.19 |
3 |
4 |
1 |
57 |
291.415 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.72 |
4.63 |
-9.95 |
2 |
4 |
0 |
53 |
290.407 |
4 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.85 |
1.39 |
-5.08 |
2 |
4 |
0 |
56 |
290.407 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.64 |
3.49 |
-34.69 |
2 |
4 |
1 |
41 |
282.367 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.64 |
3.28 |
-9.67 |
1 |
4 |
0 |
40 |
281.359 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.64 |
5.97 |
-103.84 |
3 |
4 |
2 |
42 |
283.375 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.12 |
6.54 |
-104.61 |
3 |
3 |
2 |
34 |
253.349 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.12 |
4.9 |
-7.7 |
1 |
3 |
0 |
28 |
251.333 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.12 |
6.31 |
-45.92 |
2 |
3 |
1 |
33 |
252.341 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.12 |
6.52 |
-104.62 |
3 |
3 |
2 |
34 |
253.349 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.12 |
4.88 |
-6.27 |
1 |
3 |
0 |
28 |
251.333 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.12 |
6.31 |
-45.97 |
2 |
3 |
1 |
33 |
252.341 |
0 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.78 |
2.69 |
-7.24 |
2 |
3 |
0 |
45 |
232.327 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2Y14-1-E |
P2Y Purinoceptor 14 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
810 |
0.22 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.18 |
-12.22 |
-154.65 |
7 |
20 |
-2 |
312 |
605.339 |
10 |
↓
|
|
|
|
Analogs
-
5112965
-
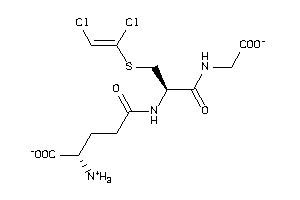
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.96 |
-2.56 |
-98.13 |
5 |
9 |
-1 |
173 |
401.248 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
-3.96 |
-3.23 |
-105.44 |
4 |
9 |
-2 |
171 |
400.24 |
11 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.29 |
2.32 |
-101.54 |
4 |
9 |
-2 |
164 |
400.24 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.15 |
-5.91 |
-138.59 |
4 |
9 |
-2 |
170 |
258.119 |
7 |
↓
|
|
Ref
Reference (pH 7)
|
-4.15 |
-7.63 |
-138.13 |
4 |
9 |
-2 |
170 |
258.119 |
7 |
↓
|
|
Mid
Mid (pH 6-8)
|
-4.15 |
-7.07 |
-49.41 |
5 |
9 |
-1 |
168 |
259.127 |
7 |
↓
|
|
|
|
Analogs
-
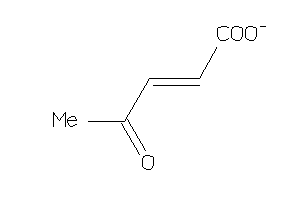
157859
-
-
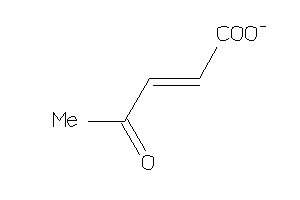
12359924
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.38 |
4.31 |
-54.71 |
0 |
3 |
-1 |
57 |
113.092 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.60 |
-4.54 |
-43.9 |
1 |
4 |
-1 |
65 |
131.086 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
-0.79 |
-1.93 |
-10.76 |
2 |
4 |
0 |
58 |
132.094 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Hi
High (pH 8-9.5)
|
-4.87 |
-2.51 |
-105.16 |
2 |
5 |
-2 |
106 |
151.143 |
3 |
↓
|
|
|
|
|
|
|
Analogs
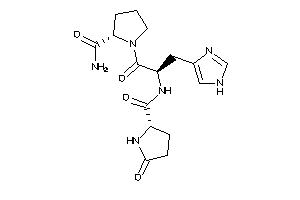
-
13523524
-
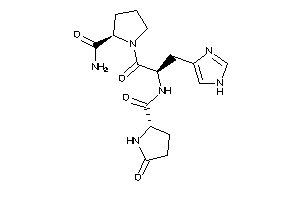
-
13523529
-
-
13523533
-
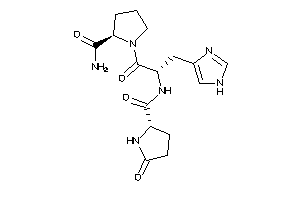
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q9R297-1-E |
Thyrotropin-releasing Hormone Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.46 |
Binding ≤ 10μM
|
|
TRFR-1-E |
Thyrotropin-releasing Hormone Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.46 |
Binding ≤ 10μM
|
|
TRFR-1-E |
Thyrotropin-releasing Hormone Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
23 |
0.41 |
Binding ≤ 10μM
|
|
TRFR-1-E |
Thyrotropin-releasing Hormone Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
42 |
0.40 |
Binding ≤ 10μM
|
|
Q9R297-1-E |
Thyrotropin-releasing Hormone Receptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.46 |
Functional ≤ 10μM
|
|
TRFR-1-E |
Thyrotropin-releasing Hormone Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3 |
0.46 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TRFR_RAT |
Q01717
|
Thyrotropin-releasing Hormone Receptor, Rat |
20 |
0.41 |
Binding ≤ 1μM
|
|
TRFR_HUMAN |
P34981
|
Thyrotropin-releasing Hormone Receptor, Human |
20 |
0.41 |
Binding ≤ 1μM
|
|
TRFR_MOUSE |
P21761
|
Thyrotropin-releasing Hormone Receptor, Mouse |
0.49 |
0.50 |
Binding ≤ 1μM
|
|
Q9R297_RAT |
Q9R297
|
Thyrotropin-releasing Hormone Receptor 2, Rat |
10 |
0.43 |
Binding ≤ 1μM
|
|
TRFR_RAT |
Q01717
|
Thyrotropin-releasing Hormone Receptor, Rat |
20 |
0.41 |
Binding ≤ 10μM
|
|
TRFR_HUMAN |
P34981
|
Thyrotropin-releasing Hormone Receptor, Human |
20 |
0.41 |
Binding ≤ 10μM
|
|
TRFR_MOUSE |
P21761
|
Thyrotropin-releasing Hormone Receptor, Mouse |
0.49 |
0.50 |
Binding ≤ 10μM
|
|
Q9R297_RAT |
Q9R297
|
Thyrotropin-releasing Hormone Receptor 2, Rat |
10 |
0.43 |
Binding ≤ 10μM
|
|
TRFR_HUMAN |
P34981
|
Thyrotropin-releasing Hormone Receptor, Human |
3 |
0.46 |
Functional ≤ 10μM
|
|
Q9R297_RAT |
Q9R297
|
Thyrotropin-releasing Hormone Receptor 2, Rat |
3 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.03 |
0.18 |
-23.59 |
5 |
10 |
0 |
150 |
362.39 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.03 |
0.01 |
-48.98 |
6 |
10 |
1 |
152 |
363.398 |
6 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.91 |
-8.71 |
-54.13 |
6 |
10 |
1 |
160 |
363.398 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.70 |
5.67 |
-47.2 |
0 |
5 |
-1 |
76 |
368.601 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.00 |
4.12 |
-43.79 |
3 |
8 |
-1 |
129 |
464.661 |
5 |
↓
|
|
|
|
Analogs
-
5339702
-
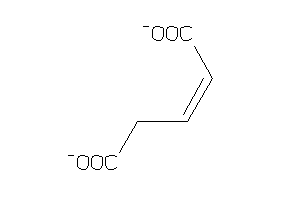
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.41 |
2.26 |
-104.21 |
0 |
4 |
-2 |
80 |
128.083 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.14 |
-1.08 |
-65.81 |
7 |
9 |
0 |
167 |
411.451 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.14 |
-1.09 |
-74.33 |
7 |
9 |
0 |
167 |
411.451 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.38 |
2.89 |
-40.21 |
3 |
7 |
1 |
97 |
318.448 |
10 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.38 |
4.37 |
-85.23 |
4 |
7 |
2 |
101 |
319.456 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.12 |
0.85 |
-59.34 |
6 |
9 |
0 |
156 |
425.478 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.12 |
0.89 |
-68.59 |
6 |
9 |
0 |
156 |
425.478 |
7 |
↓
|
|
|
|
Analogs
-
896698
-
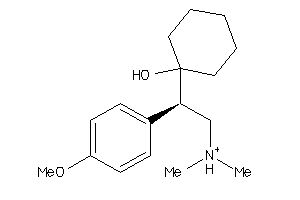
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA1A_RAT |
P43140
|
Alpha-1a Adrenergic Receptor, Rat |
109 |
0.49 |
Binding ≤ 1μM
|
|
SC6A3_RAT |
P23977
|
Dopamine Transporter, Rat |
360 |
0.45 |
Binding ≤ 1μM
|
|
SC6A2_HUMAN |
P23975
|
Norepinephrine Transporter, Human |
385 |
0.45 |
Binding ≤ 1μM
|
|
Q9WTR4_RAT |
Q9WTR4
|
Norepinephrine Transporter, Rat |
138 |
0.48 |
Binding ≤ 1μM
|
|
SC6A4_HUMAN |
P31645
|
Serotonin Transporter, Human |
27 |
0.53 |
Binding ≤ 1μM
|
|
SC6A4_RAT |
P31652
|
Serotonin Transporter, Rat |
210 |
0.47 |
Binding ≤ 1μM
|
|
Q63380_RAT |
Q63380
|
Transporter, Rat |
640 |
0.43 |
Binding ≤ 1μM
|
|
ADA1A_RAT |
P43140
|
Alpha-1a Adrenergic Receptor, Rat |
109 |
0.49 |
Binding ≤ 10μM
|
|
SC6A3_HUMAN |
Q01959
|
Dopamine Transporter, Human |
7340 |
0.36 |
Binding ≤ 10μM
|
|
SC6A3_RAT |
P23977
|
Dopamine Transporter, Rat |
360 |
0.45 |
Binding ≤ 10μM
|
|
Q9WTR4_RAT |
Q9WTR4
|
Norepinephrine Transporter, Rat |
138 |
0.48 |
Binding ≤ 10μM
|
|
SC6A2_HUMAN |
P23975
|
Norepinephrine Transporter, Human |
1060 |
0.42 |
Binding ≤ 10μM
|
|
SC6A4_RAT |
P31652
|
Serotonin Transporter, Rat |
210 |
0.47 |
Binding ≤ 10μM
|
|
SC6A4_HUMAN |
P31645
|
Serotonin Transporter, Human |
27 |
0.53 |
Binding ≤ 10μM
|
|
Q63380_RAT |
Q63380
|
Transporter, Rat |
1400 |
0.41 |
Binding ≤ 10μM
|
|
Q9WTR4_RAT |
Q9WTR4
|
Norepinephrine Transporter, Rat |
3140 |
0.39 |
Functional ≤ 10μM
|
|
SC6A4_RAT |
P31652
|
Serotonin Transporter, Rat |
100 |
0.49 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
7.36 |
-34.98 |
2 |
3 |
1 |
34 |
278.416 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.81 |
7.21 |
-3.41 |
1 |
3 |
0 |
33 |
277.408 |
5 |
↓
|
|
|
|
Analogs
-
6016
-
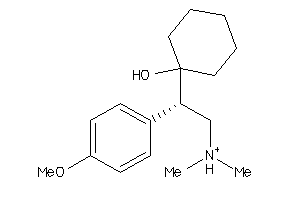
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA1A_RAT |
P43140
|
Alpha-1a Adrenergic Receptor, Rat |
109 |
0.49 |
Binding ≤ 1μM
|
|
SC6A3_RAT |
P23977
|
Dopamine Transporter, Rat |
360 |
0.45 |
Binding ≤ 1μM
|
|
SC6A2_HUMAN |
P23975
|
Norepinephrine Transporter, Human |
385 |
0.45 |
Binding ≤ 1μM
|
|
Q9WTR4_RAT |
Q9WTR4
|
Norepinephrine Transporter, Rat |
138 |
0.48 |
Binding ≤ 1μM
|
|
SC6A4_HUMAN |
P31645
|
Serotonin Transporter, Human |
27 |
0.53 |
Binding ≤ 1μM
|
|
SC6A4_RAT |
P31652
|
Serotonin Transporter, Rat |
210 |
0.47 |
Binding ≤ 1μM
|
|
Q63380_RAT |
Q63380
|
Transporter, Rat |
640 |
0.43 |
Binding ≤ 1μM
|
|
ADA1A_RAT |
P43140
|
Alpha-1a Adrenergic Receptor, Rat |
109 |
0.49 |
Binding ≤ 10μM
|
|
SC6A3_HUMAN |
Q01959
|
Dopamine Transporter, Human |
7340 |
0.36 |
Binding ≤ 10μM
|
|
SC6A3_RAT |
P23977
|
Dopamine Transporter, Rat |
360 |
0.45 |
Binding ≤ 10μM
|
|
Q9WTR4_RAT |
Q9WTR4
|
Norepinephrine Transporter, Rat |
138 |
0.48 |
Binding ≤ 10μM
|
|
SC6A2_HUMAN |
P23975
|
Norepinephrine Transporter, Human |
1060 |
0.42 |
Binding ≤ 10μM
|
|
SC6A4_RAT |
P31652
|
Serotonin Transporter, Rat |
210 |
0.47 |
Binding ≤ 10μM
|
|
SC6A4_HUMAN |
P31645
|
Serotonin Transporter, Human |
27 |
0.53 |
Binding ≤ 10μM
|
|
Q63380_RAT |
Q63380
|
Transporter, Rat |
1400 |
0.41 |
Binding ≤ 10μM
|
|
Q9WTR4_RAT |
Q9WTR4
|
Norepinephrine Transporter, Rat |
3140 |
0.39 |
Functional ≤ 10μM
|
|
SC6A4_RAT |
P31652
|
Serotonin Transporter, Rat |
100 |
0.49 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.81 |
7.34 |
-35.59 |
2 |
3 |
1 |
34 |
278.416 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.81 |
5.04 |
-4.2 |
1 |
3 |
0 |
33 |
277.408 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.04 |
-0.17 |
-39.05 |
2 |
3 |
1 |
46 |
115.156 |
1 |
↓
|
|
|
|
Analogs
-
37995315
-
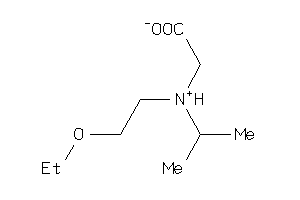
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.48 |
2.12 |
-35.34 |
1 |
4 |
0 |
53 |
145.158 |
2 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
22046874
-
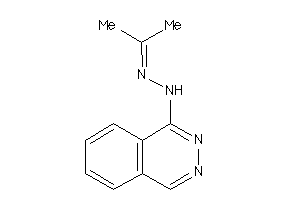
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.84 |
5.71 |
-62.47 |
1 |
6 |
-1 |
90 |
229.219 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.84 |
5.86 |
-63.06 |
2 |
6 |
0 |
92 |
230.227 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.79 |
-1.02 |
-47.36 |
2 |
2 |
1 |
21 |
305.854 |
4 |
↓
|
|