|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.85 |
-3.6 |
-11.99 |
3 |
7 |
0 |
105 |
256.258 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.85 |
-2.14 |
-17.91 |
3 |
7 |
0 |
105 |
256.258 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.85 |
-4.23 |
-13.48 |
3 |
7 |
0 |
105 |
256.258 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.85 |
-3.55 |
-13.22 |
3 |
7 |
0 |
105 |
256.258 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.51 |
9.15 |
-8.57 |
0 |
3 |
0 |
39 |
280.323 |
3 |
↓
|
|
|
|
Analogs
-
34567204
-
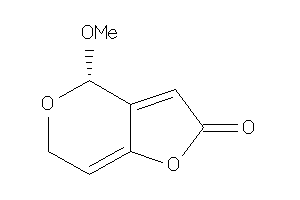
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
3800 |
0.69 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9_9HEPC |
A3EZI9
|
Hepatitis C Virus NS3 Protease/helicase, 9hepc |
3800 |
0.69 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.39 |
0.94 |
-12.15 |
1 |
4 |
0 |
60 |
154.121 |
0 |
↓
|
|
|
|
Analogs
-
34567204
-

Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9-1-V |
Hepatitis C Virus NS3 Protease/helicase (cluster #1 Of 3), Viral |
Viruses |
3800 |
0.69 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
A3EZI9_9HEPC |
A3EZI9
|
Hepatitis C Virus NS3 Protease/helicase, 9hepc |
3800 |
0.69 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.39 |
0.94 |
-12.15 |
1 |
4 |
0 |
60 |
154.121 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
10000 |
0.64 |
Binding ≤ 10μM
|
|
CAH12-4-E |
Carbonic Anhydrase XII (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
4100 |
0.69 |
Binding ≤ 10μM
|
|
CAH15-1-E |
Carbonic Anhydrase 15 (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
9230 |
0.64 |
Binding ≤ 10μM
|
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
6200 |
0.66 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
7100 |
0.66 |
Binding ≤ 10μM
|
|
CAH7-4-E |
Carbonic Anhydrase VII (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
9100 |
0.64 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.68 |
0.54 |
-10.06 |
2 |
3 |
0 |
49 |
151.165 |
1 |
↓
|
|
|
|
Analogs
-
4217364
-
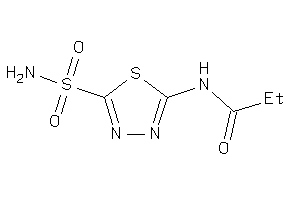
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH-1-A |
Carbonic Anhydrase (cluster #1 Of 2), Archaea |
Archaea |
60 |
0.78 |
Binding ≤ 10μM
|
|
CYNT-2-B |
Carbonic Anhydrase (cluster #2 Of 3), Bacterial |
Bacteria |
9 |
0.87 |
Binding ≤ 10μM
|
|
P96878-1-B |
PROBABLE TRANSMEMBRANE CARBONIC ANHYDRASE (CARBONATE DEHYDRATASE) (CARBONIC DEHYDRATASE) (cluster #1 Of 2), Bacterial |
Bacteria |
12 |
0.85 |
Binding ≤ 10μM
|
|
Y1284-1-B |
Uncharacterized Protein Rv1284/MT1322 (cluster #1 Of 2), Bacterial |
Bacteria |
481 |
0.68 |
Binding ≤ 10μM
|
|
B5SU02-5-E |
Alpha Carbonic Anhydrase (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
16 |
0.84 |
Binding ≤ 10μM
|
|
C0IX24-4-E |
Carbonic Anhydrase (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
74 |
0.77 |
Binding ≤ 10μM
|
|
CAH1-10-E |
Carbonic Anhydrase I (cluster #10 Of 12), Eukaryotic |
Eukaryotes |
900 |
0.65 |
Binding ≤ 10μM
|
|
CAH10-2-E |
Carbonic Anhydrase-related Protein 10 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
440 |
0.68 |
Binding ≤ 10μM
|
|
CAH11-2-E |
Carbonic Anhydrase-related Protein 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
440 |
0.68 |
Binding ≤ 10μM
|
|
CAH12-8-E |
Carbonic Anhydrase XII (cluster #8 Of 9), Eukaryotic |
Eukaryotes |
6 |
0.89 |
Binding ≤ 10μM |
|
CAH13-2-E |
Carbonic Anhydrase XIII (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
490 |
0.68 |
Binding ≤ 10μM
|
|
CAH14-7-E |
Carbonic Anhydrase XIV (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
440 |
0.68 |
Binding ≤ 10μM
|
|
CAH15-6-E |
Carbonic Anhydrase 15 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
72 |
0.77 |
Binding ≤ 10μM
|
|
CAH2-11-E |
Carbonic Anhydrase II (cluster #11 Of 15), Eukaryotic |
Eukaryotes |
7 |
0.88 |
Binding ≤ 10μM
|
|
CAH3-5-E |
Carbonic Anhydrase III (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
7 |
0.88 |
Binding ≤ 10μM
|
|
CAH4-12-E |
Carbonic Anhydrase IV (cluster #12 Of 16), Eukaryotic |
Eukaryotes |
72 |
0.77 |
Binding ≤ 10μM
|
|
CAH5A-2-E |
Carbonic Anhydrase VA (cluster #2 Of 10), Eukaryotic |
Eukaryotes |
7 |
0.88 |
Binding ≤ 10μM
|
|
CAH5B-2-E |
Carbonic Anhydrase VB (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
54 |
0.78 |
Binding ≤ 10μM |
|
CAH6-1-E |
Carbonic Anhydrase VI (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
7 |
0.88 |
Binding ≤ 10μM
|
|
CAH7-7-E |
Carbonic Anhydrase VII (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
16 |
0.84 |
Binding ≤ 10μM
|
|
CAH8-2-E |
Carbonic Anhydrase-related Protein 8 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
440 |
0.68 |
Binding ≤ 10μM
|
|
CAH9-10-E |
Carbonic Anhydrase IX (cluster #10 Of 11), Eukaryotic |
Eukaryotes |
1000 |
0.65 |
Binding ≤ 10μM
|
|
Q2PCB5-1-E |
Carbonic Anhydrase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5720 |
0.56 |
Binding ≤ 10μM |
|
CAH1-3-E |
Carbonic Anhydrase I (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
250 |
0.71 |
Functional ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.92 |
Functional ≤ 10μM
|
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
70 |
0.77 |
Functional ≤ 10μM
|
|
CAH9-1-E |
Carbonic Anhydrase IX (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
25 |
0.82 |
Functional ≤ 10μM
|
|
CAN-3-F |
Carbonic Anhydrase (cluster #3 Of 3), Fungal |
Fungi |
83 |
0.76 |
Binding ≤ 10μM
|
|
Q3I4V7-2-F |
Carbonic Anhydrase 2 (cluster #2 Of 4), Fungal |
Fungi |
10 |
0.86 |
Binding ≤ 10μM
|
|
Q5AJ71-3-F |
Carbonic Anhydrase (cluster #3 Of 4), Fungal |
Fungi |
40 |
0.80 |
Binding ≤ 10μM
|
|
Q6FTL6-1-F |
Carbonic Anhydrase (cluster #1 Of 1), Fungal |
Fungi |
11 |
0.86 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
B5SU02_9CNID |
B5SU02
|
Alpha Carbonic Anhydrase, 9cnid |
16 |
0.84 |
Binding ≤ 1μM
|
|
CAH_METTE |
P40881
|
Carbonic Anhydrase, Mette |
60 |
0.78 |
Binding ≤ 1μM
|
|
Q6FTL6_CANGA |
Q6FTL6
|
Carbonic Anhydrase, Canga |
11 |
0.86 |
Binding ≤ 1μM
|
|
Q5AJ71_CANAL |
Q5AJ71
|
Carbonic Anhydrase, Canal |
130 |
0.74 |
Binding ≤ 1μM
|
|
CYNT_MYCTU |
O53573
|
Carbonic Anhydrase, Myctu |
9 |
0.87 |
Binding ≤ 1μM
|
|
C0IX24_9CNID |
C0IX24
|
Carbonic Anhydrase, 9cnid |
74 |
0.77 |
Binding ≤ 1μM
|
|
CAN_YEAST |
P53615
|
Carbonic Anhydrase, Yeast |
82.6 |
0.76 |
Binding ≤ 1μM
|
|
CYNT_HELPY |
O24855
|
Carbonic Anhydrase 1, Helpy |
20 |
0.83 |
Binding ≤ 1μM
|
|
CAH15_MOUSE |
Q99N23
|
Carbonic Anhydrase 15, Mouse |
72 |
0.77 |
Binding ≤ 1μM
|
|
Q3I4V7_CRYNV |
Q3I4V7
|
Carbonic Anhydrase 2, Crynv |
10 |
0.86 |
Binding ≤ 1μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH2_BOVIN |
P00921
|
Carbonic Anhydrase II, Bovin |
16 |
0.84 |
Binding ≤ 1μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH2_RAT |
P27139
|
Carbonic Anhydrase II, Rat |
12 |
0.85 |
Binding ≤ 1μM
|
|
CAH3_RAT |
P14141
|
Carbonic Anhydrase III, Rat |
12 |
0.85 |
Binding ≤ 1μM
|
|
CAH3_HUMAN |
P07451
|
Carbonic Anhydrase III, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH4_RAT |
P48284
|
Carbonic Anhydrase IV, Rat |
12 |
0.85 |
Binding ≤ 1μM
|
|
CAH4_BOVIN |
Q95323
|
Carbonic Anhydrase IV, Bovin |
120 |
0.75 |
Binding ≤ 1μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH5A_RAT |
P43165
|
Carbonic Anhydrase VA, Rat |
12 |
0.85 |
Binding ≤ 1μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH5A_MOUSE |
P23589
|
Carbonic Anhydrase VA, Mouse |
58 |
0.78 |
Binding ≤ 1μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH5B_MOUSE |
Q9QZA0
|
Carbonic Anhydrase VB, Mouse |
58 |
0.78 |
Binding ≤ 1μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH6_BOVIN |
P18915
|
Carbonic Anhydrase VI, Bovin |
16 |
0.84 |
Binding ≤ 1μM
|
|
CAH7_MOUSE |
Q9ERQ8
|
Carbonic Anhydrase VII, Mouse |
16 |
0.84 |
Binding ≤ 1μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH13_HUMAN |
Q8N1Q1
|
Carbonic Anhydrase XIII, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH13_MOUSE |
Q9D6N1
|
Carbonic Anhydrase XIII, Mouse |
17 |
0.84 |
Binding ≤ 1μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH10_HUMAN |
Q9NS85
|
Carbonic Anhydrase-related Protein 10, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH11_HUMAN |
O75493
|
Carbonic Anhydrase-related Protein 2, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH8_HUMAN |
P35219
|
Carbonic Anhydrase-related Protein 8, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
P96878_MYCTU |
P96878
|
PROBABLE TRANSMEMBRANE CARBONIC ANHYDRASE (CARBONATE DEHYDRATASE) (CARBONIC DEHYDRATASE), Myctu |
100 |
0.75 |
Binding ≤ 1μM
|
|
Y1284_MYCTU |
P64797
|
Uncharacterized Protein Rv1284/MT1322, Myctu |
250 |
0.71 |
Binding ≤ 1μM
|
|
B5SU02_9CNID |
B5SU02
|
Alpha Carbonic Anhydrase, 9cnid |
16 |
0.84 |
Binding ≤ 10μM
|
|
Q2PCB5_DICLA |
Q2PCB5
|
Carbonic Anhydrase, Dicla |
5720 |
0.56 |
Binding ≤ 10μM |
|
CYNT_MYCTU |
O53573
|
Carbonic Anhydrase, Myctu |
9 |
0.87 |
Binding ≤ 10μM
|
|
C0IX24_9CNID |
C0IX24
|
Carbonic Anhydrase, 9cnid |
74 |
0.77 |
Binding ≤ 10μM
|
|
CAN_YEAST |
P53615
|
Carbonic Anhydrase, Yeast |
82.6 |
0.76 |
Binding ≤ 10μM
|
|
Q6FTL6_CANGA |
Q6FTL6
|
Carbonic Anhydrase, Canga |
11 |
0.86 |
Binding ≤ 10μM
|
|
CAH_METTE |
P40881
|
Carbonic Anhydrase, Mette |
1430 |
0.63 |
Binding ≤ 10μM
|
|
Q5AJ71_CANAL |
Q5AJ71
|
Carbonic Anhydrase, Canal |
130 |
0.74 |
Binding ≤ 10μM
|
|
CYNT_HELPY |
O24855
|
Carbonic Anhydrase 1, Helpy |
20 |
0.83 |
Binding ≤ 10μM
|
|
CAH15_MOUSE |
Q99N23
|
Carbonic Anhydrase 15, Mouse |
72 |
0.77 |
Binding ≤ 10μM
|
|
Q3I4V7_CRYNV |
Q3I4V7
|
Carbonic Anhydrase 2, Crynv |
10 |
0.86 |
Binding ≤ 10μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH2_RAT |
P27139
|
Carbonic Anhydrase II, Rat |
12 |
0.85 |
Binding ≤ 10μM
|
|
CAH2_BOVIN |
P00921
|
Carbonic Anhydrase II, Bovin |
16 |
0.84 |
Binding ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH3_RAT |
P14141
|
Carbonic Anhydrase III, Rat |
12 |
0.85 |
Binding ≤ 10μM
|
|
CAH3_HUMAN |
P07451
|
Carbonic Anhydrase III, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH4_RAT |
P48284
|
Carbonic Anhydrase IV, Rat |
12 |
0.85 |
Binding ≤ 10μM
|
|
CAH4_BOVIN |
Q95323
|
Carbonic Anhydrase IV, Bovin |
120 |
0.75 |
Binding ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH5A_MOUSE |
P23589
|
Carbonic Anhydrase VA, Mouse |
58 |
0.78 |
Binding ≤ 10μM
|
|
CAH5A_RAT |
P43165
|
Carbonic Anhydrase VA, Rat |
12 |
0.85 |
Binding ≤ 10μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH5B_MOUSE |
Q9QZA0
|
Carbonic Anhydrase VB, Mouse |
58 |
0.78 |
Binding ≤ 10μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH6_BOVIN |
P18915
|
Carbonic Anhydrase VI, Bovin |
16 |
0.84 |
Binding ≤ 10μM
|
|
CAH7_MOUSE |
Q9ERQ8
|
Carbonic Anhydrase VII, Mouse |
16 |
0.84 |
Binding ≤ 10μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH13_MOUSE |
Q9D6N1
|
Carbonic Anhydrase XIII, Mouse |
1050 |
0.64 |
Binding ≤ 10μM
|
|
CAH13_HUMAN |
Q8N1Q1
|
Carbonic Anhydrase XIII, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH10_HUMAN |
Q9NS85
|
Carbonic Anhydrase-related Protein 10, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH11_HUMAN |
O75493
|
Carbonic Anhydrase-related Protein 2, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH8_HUMAN |
P35219
|
Carbonic Anhydrase-related Protein 8, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
P96878_MYCTU |
P96878
|
PROBABLE TRANSMEMBRANE CARBONIC ANHYDRASE (CARBONATE DEHYDRATASE) (CARBONIC DEHYDRATASE), Myctu |
100 |
0.75 |
Binding ≤ 10μM
|
|
Y1284_MYCTU |
P64797
|
Uncharacterized Protein Rv1284/MT1322, Myctu |
250 |
0.71 |
Binding ≤ 10μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
250 |
0.71 |
Functional ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
12 |
0.85 |
Functional ≤ 10μM
|
|
CAH4_BOVIN |
Q95323
|
Carbonic Anhydrase IV, Bovin |
70 |
0.77 |
Functional ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
25 |
0.82 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.15 |
-4.36 |
-19.16 |
3 |
7 |
0 |
115 |
222.251 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.15 |
-3.88 |
-51.46 |
2 |
7 |
-1 |
113 |
221.243 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH2-4-E |
Carbonic Anhydrase II (cluster #4 Of 15), Eukaryotic |
Eukaryotes |
47 |
2.05 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.86 |
-1.54 |
-12.98 |
2 |
3 |
0 |
49 |
75.067 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.61 |
5.56 |
-6.8 |
1 |
2 |
0 |
37 |
204.269 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.61 |
6.34 |
-41.22 |
0 |
2 |
-1 |
40 |
203.261 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DRTS-1-E |
Bifunctional Dihydrofolate Reductase-thymidylate Synthase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8000 |
0.34 |
Binding ≤ 10μM
|
|
DYR-1-E |
Dihydrofolate Reductase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
500 |
0.42 |
Binding ≤ 10μM
|
|
KCNH2-5-E |
HERG (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
3620 |
0.36 |
Binding ≤ 10μM
|
|
B0BL08-1-B |
Dihydrofolate Reductase (cluster #1 Of 1), Bacterial |
Bacteria |
10 |
0.53 |
Binding ≤ 10μM
|
|
DYR-1-B |
Dihydrofolate Reductase (cluster #1 Of 4), Bacterial |
Bacteria |
2700 |
0.37 |
Binding ≤ 10μM
|
|
DYR1-1-B |
Dihydrofolate Reductase Type 1 (cluster #1 Of 1), Bacterial |
Bacteria |
20 |
0.51 |
Binding ≤ 10μM
|
|
O30463-1-B |
Dihydrofolate Reductase (cluster #1 Of 1), Bacterial |
Bacteria |
300 |
0.43 |
Binding ≤ 10μM
|
|
DYR-1-F |
Dihydrofolate Reductase (cluster #1 Of 1), Fungal |
Fungi |
152 |
0.45 |
Binding ≤ 10μM
|
|
Z50339-1-O |
Pneumocystis Carinii (cluster #1 Of 2), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z50362-1-O |
Lactobacillus Casei (cluster #1 Of 1), Other |
Other |
103 |
0.47 |
Functional ≤ 10μM
|
|
Z50424-1-O |
Cryptosporidium Parvum (cluster #1 Of 2), Other |
Other |
4000 |
0.36 |
Functional ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
9309 |
0.34 |
Functional ≤ 10μM
|
|
Z50472-1-O |
Toxoplasma Gondii (cluster #1 Of 4), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DYR_MOUSE |
P00375
|
Dihydrofolate Reductase, Mouse |
500 |
0.42 |
Binding ≤ 1μM
|
|
DYR_HUMAN |
P00374
|
Dihydrofolate Reductase, Human |
1 |
0.60 |
Binding ≤ 1μM
|
|
O30463_MYCAV |
O30463
|
Dihydrofolate Reductase, Mycav |
190 |
0.45 |
Binding ≤ 1μM
|
|
B0BL08_ECOLX |
B0BL08
|
Dihydrofolate Reductase, Ecolx |
10 |
0.53 |
Binding ≤ 1μM
|
|
DYR_PNECA |
P16184
|
Dihydrofolate Reductase, Pneca |
152 |
0.45 |
Binding ≤ 1μM
|
|
DRTS_PLAFK |
P13922
|
Dihydrofolate Reductase, Plafk |
10 |
0.53 |
Binding ≤ 1μM
|
|
DYR_ECOLI |
P0ABQ4
|
Dihydrofolate Reductase, Ecoli |
1.3 |
0.59 |
Binding ≤ 1μM
|
|
DYR_STAAU |
P0A017
|
Dihydrofolate Reductase, Staau |
1.24 |
0.59 |
Binding ≤ 1μM |
|
DYR_LACCA |
P00381
|
Dihydrofolate Reductase, Lacca |
620 |
0.41 |
Binding ≤ 1μM
|
|
DYR1_ECOLX |
P00382
|
Dihydrofolate Reductase Type 1, Ecolx |
10 |
0.53 |
Binding ≤ 1μM
|
|
DYR_LACCA |
P00381
|
Dihydrofolate Reductase, Lacca |
620 |
0.41 |
Binding ≤ 10μM
|
|
DYR_MOUSE |
P00375
|
Dihydrofolate Reductase, Mouse |
500 |
0.42 |
Binding ≤ 10μM
|
|
DYR_HUMAN |
P00374
|
Dihydrofolate Reductase, Human |
1 |
0.60 |
Binding ≤ 10μM
|
|
O30463_MYCAV |
O30463
|
Dihydrofolate Reductase, Mycav |
190 |
0.45 |
Binding ≤ 10μM
|
|
B0BL08_ECOLX |
B0BL08
|
Dihydrofolate Reductase, Ecolx |
10 |
0.53 |
Binding ≤ 10μM
|
|
DRTS_TOXGO |
Q07422
|
Dihydrofolate Reductase, Toxgo |
2700 |
0.37 |
Binding ≤ 10μM
|
|
DYR_PNECA |
P16184
|
Dihydrofolate Reductase, Pneca |
152 |
0.45 |
Binding ≤ 10μM
|
|
DRTS_PLAFK |
P13922
|
Dihydrofolate Reductase, Plafk |
10 |
0.53 |
Binding ≤ 10μM
|
|
DYR_ECOLI |
P0ABQ4
|
Dihydrofolate Reductase, Ecoli |
1.3 |
0.59 |
Binding ≤ 10μM
|
|
DYR_STAAU |
P0A017
|
Dihydrofolate Reductase, Staau |
1.24 |
0.59 |
Binding ≤ 10μM |
|
DYR1_ECOLX |
P00382
|
Dihydrofolate Reductase Type 1, Ecolx |
10 |
0.53 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
3620 |
0.36 |
Binding ≤ 10μM
|
|
Z50424 |
Z50424
|
Cryptosporidium Parvum |
4000 |
0.36 |
Functional ≤ 10μM
|
|
Z50362 |
Z50362
|
Lactobacillus Casei |
103 |
0.47 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
1096 |
0.40 |
Functional ≤ 10μM
|
|
Z50339 |
Z50339
|
Pneumocystis Carinii |
0.1 |
0.67 |
Functional ≤ 10μM
|
|
Z50472 |
Z50472
|
Toxoplasma Gondii |
0.1 |
0.67 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.00 |
4.4 |
-13.71 |
4 |
7 |
0 |
106 |
290.323 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.00 |
4.89 |
-42.71 |
5 |
7 |
1 |
107 |
291.331 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80211-5-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #5 Of 5), Other |
Other |
940 |
0.50 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80211 |
Z80211
|
LoVo (Colon Adenocarcinoma Cells) |
940 |
0.50 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.27 |
8.33 |
-11.82 |
0 |
3 |
0 |
43 |
228.247 |
0 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
391883
-
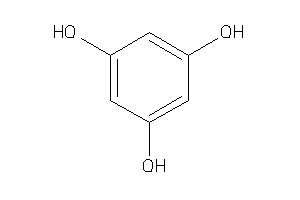
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH12-9-E |
Carbonic Anhydrase XII (cluster #9 Of 9), Eukaryotic |
Eukaryotes |
7500 |
0.90 |
Binding ≤ 10μM
|
|
CAH2-5-E |
Carbonic Anhydrase II (cluster #5 Of 15), Eukaryotic |
Eukaryotes |
7700 |
0.89 |
Binding ≤ 10μM
|
|
CAH5A-8-E |
Carbonic Anhydrase VA (cluster #8 Of 10), Eukaryotic |
Eukaryotes |
8700 |
0.89 |
Binding ≤ 10μM
|
|
CAH5B-8-E |
Carbonic Anhydrase VB (cluster #8 Of 9), Eukaryotic |
Eukaryotes |
7100 |
0.90 |
Binding ≤ 10μM
|
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
3600 |
0.95 |
Binding ≤ 10μM
|
|
Q2PCB5-2-E |
Carbonic Anhydrase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6420 |
0.91 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.96 |
-1.22 |
-5.57 |
2 |
2 |
0 |
40 |
110.112 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.32 |
1.74 |
-46.85 |
0 |
2 |
-1 |
40 |
109.104 |
0 |
↓
|
|
|
|
|
|
|
Analogs
-
2581651
-
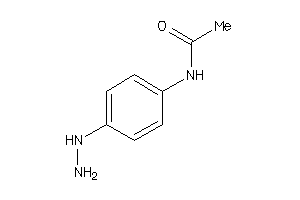
-
34396225
-
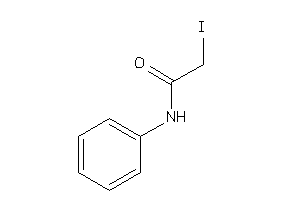
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.16 |
3.39 |
-8.79 |
1 |
2 |
0 |
29 |
135.166 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHA2-3-E |
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
180 |
0.94 |
Binding ≤ 10μM
|
|
ACHA7-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
3509 |
0.76 |
Binding ≤ 10μM
|
|
ACHB2-3-E |
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit (cluster #3 Of 7), Eukaryotic |
Eukaryotes |
180 |
0.94 |
Binding ≤ 10μM
|
|
ACHB4-4-E |
Neuronal Acetylcholine Receptor Subunit Beta-4 (cluster #4 Of 7), Eukaryotic |
Eukaryotes |
180 |
0.94 |
Binding ≤ 10μM
|
|
ACHD-2-E |
Acetylcholine Receptor Protein Delta Chain (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5000 |
0.74 |
Binding ≤ 10μM
|
|
ACHP-1-E |
Acetylcholine-binding Protein (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3162 |
0.77 |
Binding ≤ 10μM
|
|
ACM1-4-E |
Muscarinic Acetylcholine Receptor M1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
770 |
0.86 |
Binding ≤ 10μM
|
|
ACM3-4-E |
Muscarinic Acetylcholine Receptor M3 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
220 |
0.93 |
Binding ≤ 10μM
|
|
ACM5-1-E |
Muscarinic Acetylcholine Receptor M5 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
790 |
0.85 |
Binding ≤ 10μM
|
|
ACHA3-1-E |
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
8900 |
0.71 |
Functional ≤ 10μM
|
|
ACHA7-2-E |
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
4000 |
0.76 |
Functional ≤ 10μM
|
|
ACM1-1-E |
Muscarinic Acetylcholine Receptor M1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1 |
1.26 |
Functional ≤ 10μM
|
|
ACM2-2-E |
Muscarinic Acetylcholine Receptor M2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
220 |
0.93 |
Functional ≤ 10μM
|
|
ACM3-1-E |
Muscarinic Acetylcholine Receptor M3 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3 |
1.19 |
Functional ≤ 10μM
|
|
ACM4-1-E |
Muscarinic Acetylcholine Receptor M4 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
10 |
1.12 |
Functional ≤ 10μM
|
|
ACM5-2-E |
Muscarinic Acetylcholine Receptor M5 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
630 |
0.87 |
Functional ≤ 10μM
|
|
Z104281-1-O |
Neuronal Acetylcholine Receptor; Alpha3/beta2 (cluster #1 Of 2), Other |
Other |
41 |
1.03 |
Binding ≤ 10μM
|
|
Z104286-1-O |
Neuronal Acetylcholine Receptor; Alpha2/beta2 (cluster #1 Of 2), Other |
Other |
11 |
1.11 |
Binding ≤ 10μM
|
|
Z104287-2-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #2 Of 3), Other |
Other |
881 |
0.85 |
Binding ≤ 10μM
|
|
Z104289-1-O |
Neuronal Acetylcholine Receptor; Alpha4/beta4 (cluster #1 Of 2), Other |
Other |
83 |
0.99 |
Binding ≤ 10μM
|
|
Z104290-3-O |
Neuronal Acetylcholine Receptor; Alpha4/beta2 (cluster #3 Of 4), Other |
Other |
8 |
1.13 |
Binding ≤ 10μM
|
|
Z104303-2-O |
Muscarinic Acetylcholine Receptor (cluster #2 Of 7), Other |
Other |
12 |
1.11 |
Binding ≤ 10μM
|
|
Z102306-2-O |
Aorta (cluster #2 Of 6), Other |
Other |
1000 |
0.84 |
Functional ≤ 10μM |
|
Z104287-1-O |
Neuronal Acetylcholine Receptor; Alpha3/beta4 (cluster #1 Of 2), Other |
Other |
8700 |
0.71 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACHD_TORCA |
P02718
|
Acetylcholine Receptor Protein Delta Chain, Torca |
43 |
1.03 |
Binding ≤ 1μM
|
|
Z104303 |
Z104303
|
Muscarinic Acetylcholine Receptor |
12 |
1.11 |
Binding ≤ 1μM
|
|
ACM1_HUMAN |
P11229
|
Muscarinic Acetylcholine Receptor M1, Human |
770 |
0.86 |
Binding ≤ 1μM
|
|
ACM3_HUMAN |
P20309
|
Muscarinic Acetylcholine Receptor M3, Human |
220 |
0.93 |
Binding ≤ 1μM
|
|
ACM5_HUMAN |
P08912
|
Muscarinic Acetylcholine Receptor M5, Human |
790 |
0.85 |
Binding ≤ 1μM
|
|
ACHA2_RAT |
P12389
|
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit, Rat |
180 |
0.94 |
Binding ≤ 1μM
|
|
ACHB2_RAT |
P12390
|
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit, Rat |
180 |
0.94 |
Binding ≤ 1μM
|
|
ACHB4_RAT |
P12392
|
Neuronal Acetylcholine Receptor Protein Beta-4 Subunit, Rat |
180 |
0.94 |
Binding ≤ 1μM
|
|
Z104286 |
Z104286
|
Neuronal Acetylcholine Receptor; Alpha2/beta2 |
11 |
1.11 |
Binding ≤ 1μM
|
|
Z104281 |
Z104281
|
Neuronal Acetylcholine Receptor; Alpha3/beta2 |
41 |
1.03 |
Binding ≤ 1μM
|
|
Z104287 |
Z104287
|
Neuronal Acetylcholine Receptor; Alpha3/beta4 |
620 |
0.87 |
Binding ≤ 1μM
|
|
Z104290 |
Z104290
|
Neuronal Acetylcholine Receptor; Alpha4/beta2 |
37.7 |
1.04 |
Binding ≤ 1μM
|
|
Z104289 |
Z104289
|
Neuronal Acetylcholine Receptor; Alpha4/beta4 |
83 |
0.99 |
Binding ≤ 1μM
|
|
ACHD_TORCA |
P02718
|
Acetylcholine Receptor Protein Delta Chain, Torca |
43 |
1.03 |
Binding ≤ 10μM
|
|
ACHP_LYMST |
P58154
|
Acetylcholine-binding Protein, Lymst |
3162.27766 |
0.77 |
Binding ≤ 10μM
|
|
Z104303 |
Z104303
|
Muscarinic Acetylcholine Receptor |
12 |
1.11 |
Binding ≤ 10μM
|
|
ACM1_HUMAN |
P11229
|
Muscarinic Acetylcholine Receptor M1, Human |
770 |
0.86 |
Binding ≤ 10μM
|
|
ACM3_HUMAN |
P20309
|
Muscarinic Acetylcholine Receptor M3, Human |
220 |
0.93 |
Binding ≤ 10μM
|
|
ACM5_HUMAN |
P08912
|
Muscarinic Acetylcholine Receptor M5, Human |
790 |
0.85 |
Binding ≤ 10μM
|
|
ACHA2_RAT |
P12389
|
Neuronal Acetylcholine Receptor Protein Alpha-2 Subunit, Rat |
180 |
0.94 |
Binding ≤ 10μM
|
|
ACHA7_RAT |
Q05941
|
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit, Rat |
3509 |
0.76 |
Binding ≤ 10μM
|
|
ACHB2_RAT |
P12390
|
Neuronal Acetylcholine Receptor Protein Beta-2 Subunit, Rat |
180 |
0.94 |
Binding ≤ 10μM
|
|
ACHB4_RAT |
P12392
|
Neuronal Acetylcholine Receptor Protein Beta-4 Subunit, Rat |
180 |
0.94 |
Binding ≤ 10μM
|
|
Z104286 |
Z104286
|
Neuronal Acetylcholine Receptor; Alpha2/beta2 |
11 |
1.11 |
Binding ≤ 10μM
|
|
Z104281 |
Z104281
|
Neuronal Acetylcholine Receptor; Alpha3/beta2 |
41 |
1.03 |
Binding ≤ 10μM
|
|
Z104287 |
Z104287
|
Neuronal Acetylcholine Receptor; Alpha3/beta4 |
620 |
0.87 |
Binding ≤ 10μM
|
|
Z104290 |
Z104290
|
Neuronal Acetylcholine Receptor; Alpha4/beta2 |
37.7 |
1.04 |
Binding ≤ 10μM
|
|
Z104289 |
Z104289
|
Neuronal Acetylcholine Receptor; Alpha4/beta4 |
83 |
0.99 |
Binding ≤ 10μM
|
|
Z102306 |
Z102306
|
Aorta |
1000 |
0.84 |
Functional ≤ 10μM |
|
ACM1_HUMAN |
P11229
|
Muscarinic Acetylcholine Receptor M1, Human |
1.1 |
1.25 |
Functional ≤ 10μM
|
|
ACM2_HUMAN |
P08172
|
Muscarinic Acetylcholine Receptor M2, Human |
220 |
0.93 |
Functional ≤ 10μM
|
|
ACM3_HUMAN |
P20309
|
Muscarinic Acetylcholine Receptor M3, Human |
3.2 |
1.19 |
Functional ≤ 10μM
|
|
ACM4_HUMAN |
P08173
|
Muscarinic Acetylcholine Receptor M4, Human |
10 |
1.12 |
Functional ≤ 10μM
|
|
ACM5_HUMAN |
P08912
|
Muscarinic Acetylcholine Receptor M5, Human |
1.2 |
1.25 |
Functional ≤ 10μM
|
|
ACHA3_RAT |
P04757
|
Neuronal Acetylcholine Receptor Protein Alpha-3 Subunit, Rat |
8900 |
0.71 |
Functional ≤ 10μM
|
|
ACHA7_HUMAN |
P36544
|
Neuronal Acetylcholine Receptor Protein Alpha-7 Subunit, Human |
4000 |
0.76 |
Functional ≤ 10μM
|
|
Z104287 |
Z104287
|
Neuronal Acetylcholine Receptor; Alpha3/beta4 |
8700 |
0.71 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.56 |
6.42 |
-34.1 |
0 |
3 |
1 |
26 |
146.21 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AMPM-1-B |
Methionine Aminopeptidase (cluster #1 Of 3), Bacterial |
Bacteria |
472 |
0.63 |
Binding ≤ 10μM
|
|
Z50607-3-O |
Human Immunodeficiency Virus 1 (cluster #3 Of 10), Other |
Other |
4160 |
0.54 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.35 |
4.04 |
-9.14 |
1 |
3 |
0 |
42 |
201.254 |
1 |
↓
|
|
|
|
Analogs
-
1530855
-
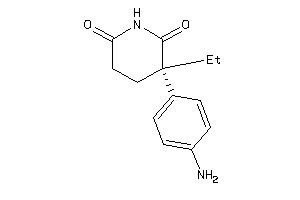
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP19A-1-E |
Cytochrome P450 19A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6200 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.76 |
2.19 |
-10.22 |
3 |
4 |
0 |
72 |
232.283 |
2 |
↓
|
|
|
|
Analogs
-
1530856
-
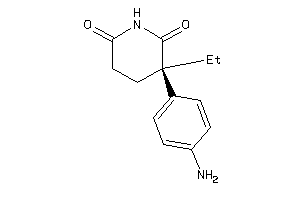
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP19A-1-E |
Cytochrome P450 19A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6200 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.76 |
2.15 |
-10.17 |
3 |
4 |
0 |
72 |
232.283 |
2 |
↓
|
|
|
|
Analogs
-
1707020
-
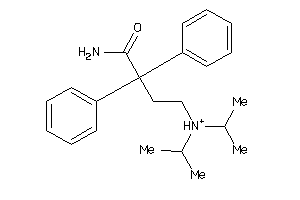
-
1718667
-
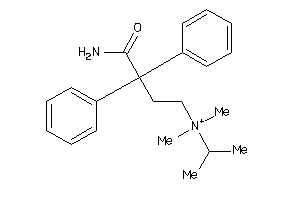
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.11 |
10 |
-37.7 |
2 |
3 |
1 |
43 |
353.53 |
8 |
↓
|
|
|
|
Analogs
-
1529820
-
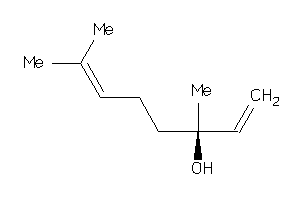
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.21 |
4.5 |
-1.93 |
1 |
1 |
0 |
20 |
154.253 |
4 |
↓
|
|
|
|
Analogs
-
1529819
-
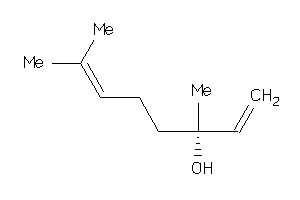
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.21 |
4.46 |
-1.88 |
1 |
1 |
0 |
20 |
154.253 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.31 |
1.79 |
-7.2 |
0 |
2 |
0 |
26 |
72.063 |
0 |
↓
|
|
|
|
Analogs
-
3874825
-
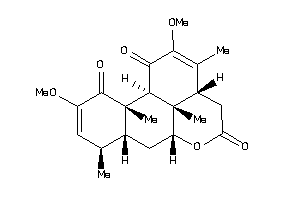
-
3874824
-
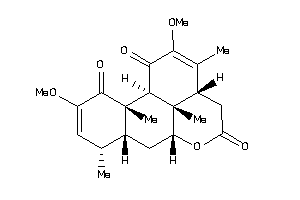
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.84 |
9.81 |
-21.35 |
0 |
6 |
0 |
79 |
388.46 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S19A2-1-E |
Thiamine Transporter 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
7600 |
0.40 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
S19A2_HUMAN |
O60779
|
Thiamine Transporter 1, Human |
7600 |
0.40 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.86 |
6.65 |
-31.15 |
2 |
4 |
1 |
56 |
243.334 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
-2.86 |
7.2 |
-87.24 |
3 |
4 |
2 |
57 |
244.342 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.95 |
2.75 |
-11.27 |
1 |
6 |
0 |
73 |
180.167 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.38 |
4.79 |
-13.99 |
2 |
3 |
0 |
58 |
226.231 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
3.48 |
2.85 |
-10.96 |
3 |
3 |
0 |
61 |
226.231 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.48 |
3.96 |
-43.55 |
2 |
3 |
-1 |
64 |
225.223 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.40 |
5.88 |
-16.84 |
0 |
3 |
0 |
27 |
188.23 |
1 |
↓
|
|
|
|
Analogs
-
40164410
-
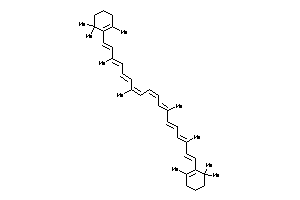
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
9.84 |
25.65 |
-2.78 |
0 |
0 |
0 |
0 |
536.888 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.03 |
4.63 |
-9.29 |
0 |
4 |
0 |
49 |
206.197 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ABCG2-1-E |
ATP-binding Cassette Sub-family G Member 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
2600 |
0.41 |
Binding ≤ 10μM
|
|
ALDR-1-E |
Aldose Reductase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
7790 |
0.38 |
Binding ≤ 10μM
|
|
AOFA-4-E |
Monoamine Oxidase A (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
1600 |
0.43 |
Binding ≤ 10μM
|
|
AOFB-4-E |
Monoamine Oxidase B (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
1600 |
0.43 |
Binding ≤ 10μM
|
|
CBR1-1-E |
Carbonyl Reductase [NADPH] 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
670 |
0.45 |
Binding ≤ 10μM
|
|
CCNB1-1-E |
G2/mitotic-specific Cyclin B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6200 |
0.38 |
Binding ≤ 10μM
|
|
CCNB2-1-E |
G2/mitotic-specific Cyclin B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6200 |
0.38 |
Binding ≤ 10μM
|
|
CCNB3-1-E |
G2/mitotic-specific Cyclin B3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
6200 |
0.38 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
6200 |
0.38 |
Binding ≤ 10μM
|
|
CDK6-1-E |
Cyclin-dependent Kinase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
6000 |
0.38 |
Binding ≤ 10μM
|
|
CP19A-3-E |
Cytochrome P450 19A1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
2600 |
0.41 |
Binding ≤ 10μM
|
|
CP1B1-1-E |
Cytochrome P450 1B1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
24 |
0.56 |
Binding ≤ 10μM
|
|
GBRA1-1-E |
GABA Receptor Alpha-1 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3020 |
0.41 |
Binding ≤ 10μM
|
|
GBRA2-1-E |
GABA Receptor Alpha-2 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3020 |
0.41 |
Binding ≤ 10μM
|
|
GBRA3-1-E |
GABA Receptor Alpha-3 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3020 |
0.41 |
Binding ≤ 10μM
|
|
GBRA4-1-E |
GABA Receptor Alpha-4 Subunit (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
3020 |
0.41 |
Binding ≤ 10μM
|
|
GBRA5-1-E |
GABA Receptor Alpha-5 Subunit (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
3020 |
0.41 |
Binding ≤ 10μM
|
|
GBRA6-6-E |
GABA Receptor Alpha-6 Subunit (cluster #6 Of 8), Eukaryotic |
Eukaryotes |
3020 |
0.41 |
Binding ≤ 10μM
|
|
GSK3A-2-E |
Glycogen Synthase Kinase-3 Alpha (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Binding ≤ 10μM
|
|
GSK3B-7-E |
Glycogen Synthase Kinase-3 Beta (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
7200 |
0.38 |
Binding ≤ 10μM
|
|
Q965D6-1-E |
3-oxoacyl-acyl-carrier Protein Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
10000 |
0.37 |
Binding ≤ 10μM
|
|
XDH-2-E |
Xanthine Dehydrogenase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
840 |
0.45 |
Binding ≤ 10μM
|
|
CP1A1-1-E |
Cytochrome P450 1A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
42 |
0.54 |
ADME/T ≤ 10μM
|
|
CP1A2-1-E |
Cytochrome P450 1A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
84 |
0.52 |
ADME/T ≤ 10μM
|
|
CP1B1-1-E |
Cytochrome P450 1B1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
16 |
0.57 |
ADME/T ≤ 10μM
|
|
CP2C9-1-E |
Cytochrome P450 2C9 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
5225 |
0.39 |
ADME/T ≤ 10μM
|
|
Z104294-2-O |
Cyclin-dependent Kinase 5/CDK5 Activator 1 (cluster #2 Of 2), Other |
Other |
3100 |
0.41 |
Binding ≤ 10μM
|
|
Z104301-4-O |
GABA-A Receptor; Anion Channel (cluster #4 Of 8), Other |
Other |
920 |
0.44 |
Binding ≤ 10μM
|
|
Z50607-3-O |
Human Immunodeficiency Virus 1 (cluster #3 Of 10), Other |
Other |
5000 |
0.39 |
Functional ≤ 10μM |
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
3100 |
0.41 |
Functional ≤ 10μM
|
|
Z80470-1-O |
SGC-7901 (Gastric Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
5800 |
0.39 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CBR1_HUMAN |
P16152
|
Carbonyl Reductase [NADPH] 1, Human |
110 |
0.51 |
Binding ≤ 1μM
|
|
CP19A_HUMAN |
P11511
|
Cytochrome P450 19A1, Human |
1 |
0.66 |
Binding ≤ 1μM
|
|
CP1B1_HUMAN |
Q16678
|
Cytochrome P450 1B1, Human |
24 |
0.56 |
Binding ≤ 1μM
|
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
920 |
0.44 |
Binding ≤ 1μM
|
|
XDH_HUMAN |
P47989
|
Xanthine Dehydrogenase, Human |
840 |
0.45 |
Binding ≤ 1μM
|
|
Q965D6_PLAFA |
Q965D6
|
3-oxoacyl-acyl-carrier Protein Reductase, Plafa |
10000 |
0.37 |
Binding ≤ 10μM
|
|
ALDR_BOVIN |
P16116
|
Aldose Reductase, Bovin |
7790 |
0.38 |
Binding ≤ 10μM
|
|
ALDR_HUMAN |
P15121
|
Aldose Reductase, Human |
7780 |
0.38 |
Binding ≤ 10μM
|
|
ABCG2_HUMAN |
Q9UNQ0
|
ATP-binding Cassette Sub-family G Member 2, Human |
1500 |
0.43 |
Binding ≤ 10μM
|
|
CBR1_HUMAN |
P16152
|
Carbonyl Reductase [NADPH] 1, Human |
110 |
0.51 |
Binding ≤ 10μM
|
|
CDK1_HUMAN |
P06493
|
Cyclin-dependent Kinase 1, Human |
6200 |
0.38 |
Binding ≤ 10μM
|
|
Z104294 |
Z104294
|
Cyclin-dependent Kinase 5/CDK5 Activator 1 |
3100 |
0.41 |
Binding ≤ 10μM
|
|
CDK6_HUMAN |
Q00534
|
Cyclin-dependent Kinase 6, Human |
6000 |
0.38 |
Binding ≤ 10μM
|
|
CP19A_HUMAN |
P11511
|
Cytochrome P450 19A1, Human |
1 |
0.66 |
Binding ≤ 10μM
|
|
CP1B1_HUMAN |
Q16678
|
Cytochrome P450 1B1, Human |
24 |
0.56 |
Binding ≤ 10μM
|
|
CCNB1_HUMAN |
P14635
|
G2/mitotic-specific Cyclin B1, Human |
6200 |
0.38 |
Binding ≤ 10μM
|
|
CCNB2_HUMAN |
O95067
|
G2/mitotic-specific Cyclin B2, Human |
6200 |
0.38 |
Binding ≤ 10μM
|
|
CCNB3_HUMAN |
Q8WWL7
|
G2/mitotic-specific Cyclin B3, Human |
6200 |
0.38 |
Binding ≤ 10μM
|
|
GBRA1_HUMAN |
P14867
|
GABA Receptor Alpha-1 Subunit, Human |
3019.95172 |
0.41 |
Binding ≤ 10μM
|
|
GBRA2_HUMAN |
P47869
|
GABA Receptor Alpha-2 Subunit, Human |
3019.95172 |
0.41 |
Binding ≤ 10μM
|
|
GBRA3_HUMAN |
P34903
|
GABA Receptor Alpha-3 Subunit, Human |
3019.95172 |
0.41 |
Binding ≤ 10μM
|
|
GBRA4_HUMAN |
P48169
|
GABA Receptor Alpha-4 Subunit, Human |
3019.95172 |
0.41 |
Binding ≤ 10μM
|
|
GBRA5_HUMAN |
P31644
|
GABA Receptor Alpha-5 Subunit, Human |
3019.95172 |
0.41 |
Binding ≤ 10μM
|
|
GBRA6_HUMAN |
Q16445
|
GABA Receptor Alpha-6 Subunit, Human |
3019.95172 |
0.41 |
Binding ≤ 10μM
|
|
Z104301 |
Z104301
|
GABA-A Receptor; Anion Channel |
920 |
0.44 |
Binding ≤ 10μM
|
|
GSK3A_HUMAN |
P49840
|
Glycogen Synthase Kinase-3 Alpha, Human |
7200 |
0.38 |
Binding ≤ 10μM
|
|
GSK3B_HUMAN |
P49841
|
Glycogen Synthase Kinase-3 Beta, Human |
7200 |
0.38 |
Binding ≤ 10μM
|
|
AOFA_HUMAN |
P21397
|
Monoamine Oxidase A, Human |
1600 |
0.43 |
Binding ≤ 10μM
|
|
AOFB_HUMAN |
P27338
|
Monoamine Oxidase B, Human |
1600 |
0.43 |
Binding ≤ 10μM
|
|
XDH_HUMAN |
P47989
|
Xanthine Dehydrogenase, Human |
840 |
0.45 |
Binding ≤ 10μM
|
|
Z80166 |
Z80166
|
HT-29 (Colon Adenocarcinoma Cells) |
3100 |
0.41 |
Functional ≤ 10μM
|
|
Z50607 |
Z50607
|
Human Immunodeficiency Virus 1 |
5000 |
0.39 |
Functional ≤ 10μM
|
|
Z80470 |
Z80470
|
SGC-7901 (Gastric Carcinoma Cells) |
5800 |
0.39 |
Functional ≤ 10μM
|
|
CP1A1_HUMAN |
P04798
|
Cytochrome P450 1A1, Human |
153 |
0.50 |
ADME/T ≤ 10μM
|
|
CP1A2_HUMAN |
P05177
|
Cytochrome P450 1A2, Human |
54 |
0.54 |
ADME/T ≤ 10μM
|
|
CP1B1_HUMAN |
Q16678
|
Cytochrome P450 1B1, Human |
16 |
0.57 |
ADME/T ≤ 10μM
|
|
CP2C9_HUMAN |
P11712
|
Cytochrome P450 2C9, Human |
5225 |
0.39 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.94 |
3.96 |
-11.62 |
2 |
4 |
0 |
71 |
254.241 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.94 |
4.94 |
-56.85 |
1 |
4 |
-1 |
73 |
253.233 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.20 |
4.26 |
-45.71 |
1 |
4 |
-1 |
73 |
253.233 |
1 |
↓
|
|
|
|
Analogs
-
3977873
-
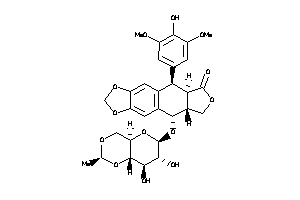
-
3977874
-
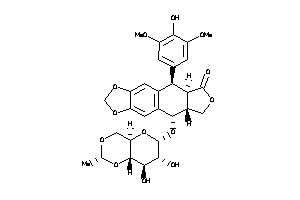
-
3977875
-
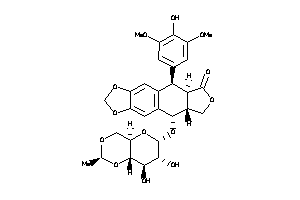
-
4245621
-
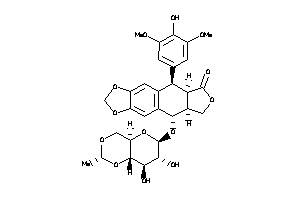
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CASP3-1-E |
Caspase-3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.20 |
Binding ≤ 10μM
|
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
810 |
0.20 |
Binding ≤ 10μM |
|
TOP2B-1-E |
DNA Topoisomerase II Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
810 |
0.20 |
Binding ≤ 10μM |
|
TOP1-1-E |
DNA Topoisomerase I (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.25 |
Functional ≤ 10μM
|
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.25 |
Functional ≤ 10μM
|
|
TOP2B-1-E |
DNA Topoisomerase II Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.25 |
Functional ≤ 10μM
|
|
Z100733-1-O |
CT26 (Colon Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
200 |
0.22 |
Functional ≤ 10μM
|
|
Z100752-1-O |
Renca (Renal Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
2900 |
0.18 |
Functional ≤ 10μM
|
|
Z103203-1-O |
A375 (cluster #1 Of 3), Other |
Other |
3920 |
0.18 |
Functional ≤ 10μM
|
|
Z103204-2-O |
A427 (cluster #2 Of 4), Other |
Other |
130 |
0.23 |
Functional ≤ 10μM
|
|
Z103413-1-O |
SW948 (cluster #1 Of 1), Other |
Other |
1300 |
0.20 |
Functional ≤ 10μM
|
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
5012 |
0.18 |
Functional ≤ 10μM
|
|
Z50587-5-O |
Homo Sapiens (cluster #5 Of 9), Other |
Other |
990 |
0.20 |
Functional ≤ 10μM
|
|
Z80008-1-O |
5637 (Epithelial Bladder Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
530 |
0.21 |
Functional ≤ 10μM
|
|
Z80025-1-O |
ACHN (Renal Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
2150 |
0.19 |
Functional ≤ 10μM
|
|
Z80026-1-O |
AGS (Gastric Adenocarcinoma Cells) (cluster #1 Of 2), Other |
Other |
360 |
0.21 |
Functional ≤ 10μM |
|
Z80035-1-O |
B16 (Melanoma Cells) (cluster #1 Of 7), Other |
Other |
9000 |
0.17 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
2 |
0.29 |
Functional ≤ 10μM
|
|
Z80064-1-O |
CCRF-CEM (T-cell Leukemia) (cluster #1 Of 9), Other |
Other |
1200 |
0.20 |
Functional ≤ 10μM
|
|
Z80068-1-O |
CCRF-SB (Lymphoblastic Leukemia Cells) (cluster #1 Of 5), Other |
Other |
100 |
0.23 |
Functional ≤ 10μM
|
|
Z80088-1-O |
CHO (Ovarian Cells) (cluster #1 Of 4), Other |
Other |
350 |
0.22 |
Functional ≤ 10μM
|
|
Z80112-1-O |
DAN-G (cluster #1 Of 2), Other |
Other |
380 |
0.21 |
Functional ≤ 10μM
|
|
Z80125-1-O |
DU-145 (Prostate Carcinoma) (cluster #1 Of 9), Other |
Other |
5160 |
0.18 |
Functional ≤ 10μM
|
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
970 |
0.20 |
Functional ≤ 10μM
|
|
Z80164-1-O |
HT-1080 (Fibrosarcoma Cells) (cluster #1 Of 6), Other |
Other |
750 |
0.20 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
6600 |
0.17 |
Functional ≤ 10μM
|
|
Z80179-1-O |
J82 (Bladder Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
800 |
0.20 |
Functional ≤ 10μM |
|
Z80186-9-O |
K562 (Erythroleukemia Cells) (cluster #9 Of 11), Other |
Other |
522 |
0.21 |
Functional ≤ 10μM
|
|
Z80188-1-O |
KB 3-1 (Cervical Epithelial Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
567 |
0.21 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
834 |
0.20 |
Functional ≤ 10μM
|
|
Z80211-1-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #1 Of 5), Other |
Other |
2300 |
0.19 |
Functional ≤ 10μM
|
|
Z80224-1-O |
MCF7 (Breast Carcinoma Cells) (cluster #1 Of 14), Other |
Other |
878 |
0.20 |
Functional ≤ 10μM
|
|
Z80244-4-O |
MDA-MB-468 (Breast Adenocarcinoma) (cluster #4 Of 7), Other |
Other |
600 |
0.21 |
Functional ≤ 10μM
|
|
Z80284-3-O |
MOLT-3 (T-lymphoblastic Leukemia Cells) (cluster #3 Of 3), Other |
Other |
65 |
0.24 |
Functional ≤ 10μM
|
|
Z80285-1-O |
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) (cluster #1 Of 4), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM |
|
Z80288-1-O |
MOVP-3 (cluster #1 Of 1), Other |
Other |
7550 |
0.17 |
Functional ≤ 10μM
|
|
Z80295-6-O |
MT4 (Lymphocytes) (cluster #6 Of 8), Other |
Other |
1250 |
0.20 |
Functional ≤ 10μM
|
|
Z80322-2-O |
NCI-H358 (Lung Carcinama Cells) (cluster #2 Of 2), Other |
Other |
1500 |
0.19 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
50 |
0.24 |
Functional ≤ 10μM
|
|
Z80390-2-O |
PC-3 (Prostate Carcinoma Cells) (cluster #2 Of 10), Other |
Other |
7800 |
0.17 |
Functional ≤ 10μM
|
|
Z80435-1-O |
RT-112 (cluster #1 Of 1), Other |
Other |
220 |
0.22 |
Functional ≤ 10μM
|
|
Z80436-1-O |
RT-4 (cluster #1 Of 1), Other |
Other |
1330 |
0.20 |
Functional ≤ 10μM
|
|
Z80473-1-O |
SISO (cluster #1 Of 2), Other |
Other |
150 |
0.23 |
Functional ≤ 10μM
|
|
Z80475-1-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #1 Of 3), Other |
Other |
300 |
0.22 |
Functional ≤ 10μM
|
|
Z80482-1-O |
SK-MEL-2 (Melanoma Cells) (cluster #1 Of 4), Other |
Other |
5720 |
0.17 |
Functional ≤ 10μM
|
|
Z80485-1-O |
SK-MEL-28 (Melanoma Cells) (cluster #1 Of 6), Other |
Other |
850 |
0.20 |
Functional ≤ 10μM
|
|
Z80490-3-O |
SK-MES-1 (cluster #3 Of 4), Other |
Other |
2500 |
0.19 |
Functional ≤ 10μM
|
|
Z80491-2-O |
SK-N-MC (Neuroepithelioma Cells) (cluster #2 Of 4), Other |
Other |
275 |
0.22 |
Functional ≤ 10μM
|
|
Z80493-2-O |
SK-OV-3 (Ovarian Carcinoma Cells) (cluster #2 Of 6), Other |
Other |
4900 |
0.18 |
Functional ≤ 10μM
|
|
Z80546-1-O |
TERT-RPE1 (Retinal Pigmented Epithelial Cells) (cluster #1 Of 1), Other |
Other |
90 |
0.23 |
Functional ≤ 10μM
|
|
Z80548-4-O |
THP-1 (Acute Monocytic Leukemia Cells) (cluster #4 Of 5), Other |
Other |
2160 |
0.19 |
Functional ≤ 10μM
|
|
Z80559-1-O |
U251 (cluster #1 Of 3), Other |
Other |
500 |
0.21 |
Functional ≤ 10μM
|
|
Z80600-2-O |
XF498 (Glioma Cells) (cluster #2 Of 2), Other |
Other |
3480 |
0.18 |
Functional ≤ 10μM
|
|
Z80601-1-O |
XRS6 (cluster #1 Of 1), Other |
Other |
140 |
0.23 |
Functional ≤ 10μM
|
|
Z80604-1-O |
YAPC (cluster #1 Of 2), Other |
Other |
1320 |
0.20 |
Functional ≤ 10μM
|
|
Z80608-1-O |
ZR-75-1 (Breast Carcinoma Cells) (cluster #1 Of 4), Other |
Other |
240 |
0.22 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
9980 |
0.17 |
Functional ≤ 10μM
|
|
Z80712-2-O |
T47D (Breast Carcinoma Cells) (cluster #2 Of 7), Other |
Other |
160 |
0.23 |
Functional ≤ 10μM
|
|
Z80819-1-O |
J774.2 (cluster #1 Of 1), Other |
Other |
3667 |
0.18 |
Functional ≤ 10μM |
|
Z80846-1-O |
F460pv8/eto Cell Line (cluster #1 Of 2), Other |
Other |
740 |
0.20 |
Functional ≤ 10μM
|
|
Z80852-1-O |
A-431 (Epidermoid Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
591 |
0.21 |
Functional ≤ 10μM
|
|
Z80866-1-O |
GLC4 Cell Line (cluster #1 Of 1), Other |
Other |
820 |
0.20 |
Functional ≤ 10μM
|
|
Z80889-1-O |
NCI-H322M (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
1590 |
0.19 |
Functional ≤ 10μM
|
|
Z80896-1-O |
NCI-H69 (Small Cell Lung Carcinoma Cells) (cluster #1 Of 1), Other |
Other |
1192 |
0.20 |
Functional ≤ 10μM
|
|
Z80901-1-O |
HaCaT (Keratinocytes) (cluster #1 Of 2), Other |
Other |
550 |
0.21 |
Functional ≤ 10μM
|
|
Z80928-1-O |
HCT-116 (Colon Carcinoma Cells) (cluster #1 Of 9), Other |
Other |
9500 |
0.17 |
Functional ≤ 10μM
|
|
Z81020-3-O |
HepG2 (Hepatoblastoma Cells) (cluster #3 Of 8), Other |
Other |
4900 |
0.18 |
Functional ≤ 10μM
|
|
Z81024-1-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #1 Of 8), Other |
Other |
90 |
0.23 |
Functional ≤ 10μM
|
|
Z81043-1-O |
N592 (cluster #1 Of 2), Other |
Other |
500 |
0.21 |
Functional ≤ 10μM
|
|
Z81054-1-O |
SF-268 (Glioblastoma Cells) (cluster #1 Of 3), Other |
Other |
3250 |
0.18 |
Functional ≤ 10μM
|
|
Z81065-1-O |
IGROV-1 (Ovarian Adenocarcinoma Cells) (cluster #1 Of 3), Other |
Other |
985 |
0.20 |
Functional ≤ 10μM
|
|
Z81072-1-O |
Jurkat (Acute Leukemic T-cells) (cluster #1 Of 10), Other |
Other |
160 |
0.23 |
Functional ≤ 10μM
|
|
Z81115-1-O |
KB (Squamous Cell Carcinoma) (cluster #1 Of 6), Other |
Other |
800 |
0.20 |
Functional ≤ 10μM
|
|
Z81126-1-O |
KYSE-150 Cell Line (cluster #1 Of 1), Other |
Other |
700 |
0.21 |
Functional ≤ 10μM
|
|
Z81128-1-O |
KYSE-520 Cell Line (cluster #1 Of 1), Other |
Other |
410 |
0.21 |
Functional ≤ 10μM
|
|
Z81130-1-O |
KYSE-70 Cell Line (cluster #1 Of 1), Other |
Other |
790 |
0.20 |
Functional ≤ 10μM
|
|
Z81160-1-O |
Lewis Lung Carcinoma Cell Line (cluster #1 Of 1), Other |
Other |
180 |
0.22 |
Functional ≤ 10μM
|
|
Z81244-1-O |
J774 (Macrophage Cells) (cluster #1 Of 1), Other |
Other |
577 |
0.21 |
Functional ≤ 10μM |
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
8930 |
0.17 |
Functional ≤ 10μM
|
|
Z81252-3-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #3 Of 11), Other |
Other |
700 |
0.21 |
Functional ≤ 10μM
|
|
Z81280-1-O |
NCI-H226 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
240 |
0.22 |
Functional ≤ 10μM
|
|
Z81284-1-O |
NCI-H647 (cluster #1 Of 2), Other |
Other |
80 |
0.24 |
Functional ≤ 10μM
|
|
Z81335-2-O |
HCT-15 (Colon Adenocarcinoma Cells) (cluster #2 Of 5), Other |
Other |
900 |
0.20 |
Functional ≤ 10μM
|
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 4), Other |
Other |
300 |
0.22 |
ADME/T ≤ 10μM
|
|
Z80291-1-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #1 Of 3), Other |
Other |
3930 |
0.18 |
ADME/T ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CASP3_HUMAN |
P42574
|
Caspase-3, Human |
1000 |
0.20 |
Binding ≤ 1μM
|
|
TOP2A_HUMAN |
P11388
|
DNA Topoisomerase II Alpha, Human |
810 |
0.20 |
Binding ≤ 1μM
|
|
TOP2B_HUMAN |
Q02880
|
DNA Topoisomerase II Beta, Human |
810 |
0.20 |
Binding ≤ 1μM
|
|
CASP3_HUMAN |
P42574
|
Caspase-3, Human |
1000 |
0.20 |
Binding ≤ 10μM
|
|
TOP2A_HUMAN |
P11388
|
DNA Topoisomerase II Alpha, Human |
810 |
0.20 |
Binding ≤ 10μM
|
|
TOP2B_HUMAN |
Q02880
|
DNA Topoisomerase II Beta, Human |
810 |
0.20 |
Binding ≤ 10μM
|
|
Z80008 |
Z80008
|
5637 (Epithelial Bladder Carcinoma Cells) |
530 |
0.21 |
Functional ≤ 10μM
|
|
Z80852 |
Z80852
|
A-431 (Epidermoid Carcinoma Cells) |
591 |
0.21 |
Functional ≤ 10μM
|
|
Z103203 |
Z103203
|
A375 |
3920 |
0.18 |
Functional ≤ 10μM
|
|
Z103204 |
Z103204
|
A427 |
130 |
0.23 |
Functional ≤ 10μM
|
|
Z80682 |
Z80682
|
A549 (Lung Carcinoma Cells) |
1060 |
0.20 |
Functional ≤ 10μM
|
|
Z80025 |
Z80025
|
ACHN (Renal Adenocarcinoma Cells) |
2150 |
0.19 |
Functional ≤ 10μM
|
|
Z80026 |
Z80026
|
AGS (Gastric Adenocarcinoma Cells) |
360 |
0.21 |
Functional ≤ 10μM
|
|
Z80035 |
Z80035
|
B16 (Melanoma Cells) |
130 |
0.23 |
Functional ≤ 10μM
|
|
Z80064 |
Z80064
|
CCRF-CEM (T-cell Leukemia) |
2 |
0.29 |
Functional ≤ 10μM
|
|
Z80068 |
Z80068
|
CCRF-SB (Lymphoblastic Leukemia Cells) |
100 |
0.23 |
Functional ≤ 10μM
|
|
Z80088 |
Z80088
|
CHO (Ovarian Cells) |
350 |
0.22 |
Functional ≤ 10μM
|
|
Z100733 |
Z100733
|
CT26 (Colon Carcinoma Cells) |
200 |
0.22 |
Functional ≤ 10μM
|
|
Z80112 |
Z80112
|
DAN-G |
380 |
0.21 |
Functional ≤ 10μM
|
|
TOP1_MOUSE |
Q04750
|
DNA Topoisomerase I, Mouse |
180 |
0.22 |
Functional ≤ 10μM
|
|
TOP1_HUMAN |
P11387
|
DNA Topoisomerase I, Human |
160 |
0.23 |
Functional ≤ 10μM
|
|
TOP2A_MOUSE |
Q01320
|
DNA Topoisomerase II Alpha, Mouse |
180 |
0.22 |
Functional ≤ 10μM
|
|
TOP2A_HUMAN |
P11388
|
DNA Topoisomerase II Alpha, Human |
160 |
0.23 |
Functional ≤ 10μM
|
|
TOP2B_MOUSE |
Q64511
|
DNA Topoisomerase II Beta, Mouse |
180 |
0.22 |
Functional ≤ 10μM
|
|
TOP2B_HUMAN |
Q02880
|
DNA Topoisomerase II Beta, Human |
160 |
0.23 |
Functional ≤ 10μM
|
|
Z80125 |
Z80125
|
DU-145 (Prostate Carcinoma) |
1300 |
0.20 |
Functional ≤ 10μM
|
|
Z80846 |
Z80846
|
F460pv8/eto Cell Line |
740 |
0.20 |
Functional ≤ 10μM
|
|
Z80866 |
Z80866
|
GLC4 Cell Line |
4000 |
0.18 |
Functional ≤ 10μM
|
|
Z80901 |
Z80901
|
HaCaT (Keratinocytes) |
550 |
0.21 |
Functional ≤ 10μM
|
|
Z80928 |
Z80928
|
HCT-116 (Colon Carcinoma Cells) |
1700 |
0.19 |
Functional ≤ 10μM
|
|
Z81335 |
Z81335
|
HCT-15 (Colon Adenocarcinoma Cells) |
1090 |
0.20 |
Functional ≤ 10μM
|
|
Z81247 |
Z81247
|
HeLa (Cervical Adenocarcinoma Cells) |
1290 |
0.20 |
Functional ≤ 10μM
|
|
Z81020 |
Z81020
|
HepG2 (Hepatoblastoma Cells) |
4100 |
0.18 |
Functional ≤ 10μM
|
|
Z80156 |
Z80156
|
HL-60 (Promyeloblast Leukemia Cells) |
0.1 |
0.33 |
Functional ≤ 10μM
|
|
Z50587 |
Z50587
|
Homo Sapiens |
990 |
0.20 |
Functional ≤ 10μM
|
|
Z80164 |
Z80164
|
HT-1080 (Fibrosarcoma Cells) |
250 |
0.22 |
Functional ≤ 10μM
|
|
Z80166 |
Z80166
|
HT-29 (Colon Adenocarcinoma Cells) |
10000 |
0.17 |
Functional ≤ 10μM
|
|
Z81065 |
Z81065
|
IGROV-1 (Ovarian Adenocarcinoma Cells) |
984.5 |
0.20 |
Functional ≤ 10μM
|
|
Z81244 |
Z81244
|
J774 (Macrophage Cells) |
576.7 |
0.21 |
Functional ≤ 10μM
|
|
Z80819 |
Z80819
|
J774.2 |
2133.3 |
0.19 |
Functional ≤ 10μM
|
|
Z80179 |
Z80179
|
J82 (Bladder Carcinoma Cells) |
2500 |
0.19 |
Functional ≤ 10μM
|
|
Z81072 |
Z81072
|
Jurkat (Acute Leukemic T-cells) |
1200 |
0.20 |
Functional ≤ 10μM
|
|
Z80186 |
Z80186
|
K562 (Erythroleukemia Cells) |
1210 |
0.20 |
Functional ≤ 10μM
|
|
Z81115 |
Z81115
|
KB (Squamous Cell Carcinoma) |
10000 |
0.17 |
Functional ≤ 10μM
|
|
Z80188 |
Z80188
|
KB 3-1 (Cervical Epithelial Carcinoma Cells) |
566.5 |
0.21 |
Functional ≤ 10μM
|
|
Z81126 |
Z81126
|
KYSE-150 Cell Line |
700 |
0.21 |
Functional ≤ 10μM
|
|
Z81128 |
Z81128
|
KYSE-520 Cell Line |
410 |
0.21 |
Functional ≤ 10μM
|
|
Z81130 |
Z81130
|
KYSE-70 Cell Line |
790 |
0.20 |
Functional ≤ 10μM
|
|
Z80193 |
Z80193
|
L1210 (Lymphocytic Leukemia Cells) |
200 |
0.22 |
Functional ≤ 10μM
|
|
Z81160 |
Z81160
|
Lewis Lung Carcinoma Cell Line |
1100 |
0.20 |
Functional ≤ 10μM
|
|
Z80211 |
Z80211
|
LoVo (Colon Adenocarcinoma Cells) |
2300 |
0.19 |
Functional ≤ 10μM
|
|
Z80224 |
Z80224
|
MCF7 (Breast Carcinoma Cells) |
1000 |
0.20 |
Functional ≤ 10μM
|
|
Z81252 |
Z81252
|
MDA-MB-231 (Breast Adenocarcinoma Cells) |
1230 |
0.20 |
Functional ≤ 10μM
|
|
Z80244 |
Z80244
|
MDA-MB-468 (Breast Adenocarcinoma) |
600 |
0.21 |
Functional ≤ 10μM
|
|
Z80284 |
Z80284
|
MOLT-3 (T-lymphoblastic Leukemia Cells) |
65 |
0.24 |
Functional ≤ 10μM
|
|
Z80285 |
Z80285
|
MOLT-4 (Acute T-lymphoblastic Leukemia Cells) |
0.1 |
0.33 |
Functional ≤ 10μM
|
|
Z80288 |
Z80288
|
MOVP-3 |
7550 |
0.17 |
Functional ≤ 10μM
|
|
Z80295 |
Z80295
|
MT4 (Lymphocytes) |
1250 |
0.20 |
Functional ≤ 10μM
|
|
Z81043 |
Z81043
|
N592 |
500 |
0.21 |
Functional ≤ 10μM
|
|
Z81280 |
Z81280
|
NCI-H226 (Non-small Cell Lung Carcinoma Cells) |
240 |
0.22 |
Functional ≤ 10μM
|
|
Z80889 |
Z80889
|
NCI-H322M (Non-small Cell Lung Carcinoma Cells) |
1590 |
0.19 |
Functional ≤ 10μM
|
|
Z80322 |
Z80322
|
NCI-H358 (Lung Carcinama Cells) |
1500 |
0.19 |
Functional ≤ 10μM
|
|
Z81024 |
Z81024
|
NCI-H460 (Non-small Cell Lung Carcinoma) |
100 |
0.23 |
Functional ≤ 10μM
|
|
Z81284 |
Z81284
|
NCI-H647 |
80 |
0.24 |
Functional ≤ 10μM
|
|
Z80896 |
Z80896
|
NCI-H69 (Small Cell Lung Carcinoma Cells) |
1192.2 |
0.20 |
Functional ≤ 10μM
|
|
Z80362 |
Z80362
|
P388 (Lymphoma Cells) |
100 |
0.23 |
Functional ≤ 10μM
|
|
Z80390 |
Z80390
|
PC-3 (Prostate Carcinoma Cells) |
1700 |
0.19 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
3981.07171 |
0.18 |
Functional ≤ 10μM
|
|
Z100752 |
Z100752
|
Renca (Renal Carcinoma Cells) |
2900 |
0.18 |
Functional ≤ 10μM
|
|
Z80435 |
Z80435
|
RT-112 |
220 |
0.22 |
Functional ≤ 10μM
|
|
Z80436 |
Z80436
|
RT-4 |
1330 |
0.20 |
Functional ≤ 10μM
|
|
Z81054 |
Z81054
|
SF-268 (Glioblastoma Cells) |
3250 |
0.18 |
Functional ≤ 10μM
|
|
Z80473 |
Z80473
|
SISO |
150 |
0.23 |
Functional ≤ 10μM
|
|
Z80475 |
Z80475
|
SK-BR-3 (Breast Adenocarcinoma) |
300 |
0.22 |
Functional ≤ 10μM
|
|
Z80482 |
Z80482
|
SK-MEL-2 (Melanoma Cells) |
1170 |
0.20 |
Functional ≤ 10μM
|
|
Z80485 |
Z80485
|
SK-MEL-28 (Melanoma Cells) |
850 |
0.20 |
Functional ≤ 10μM
|
|
Z80490 |
Z80490
|
SK-MES-1 |
2500 |
0.19 |
Functional ≤ 10μM
|
|
Z80491 |
Z80491
|
SK-N-MC (Neuroepithelioma Cells) |
275 |
0.22 |
Functional ≤ 10μM
|
|
Z80493 |
Z80493
|
SK-OV-3 (Ovarian Carcinoma Cells) |
1720 |
0.19 |
Functional ≤ 10μM
|
|
Z103413 |
Z103413
|
SW948 |
1300 |
0.20 |
Functional ≤ 10μM
|
|
Z80712 |
Z80712
|
T47D (Breast Carcinoma Cells) |
100 |
0.23 |
Functional ≤ 10μM
|
|
Z80546 |
Z80546
|
TERT-RPE1 (Retinal Pigmented Epithelial Cells) |
90 |
0.23 |
Functional ≤ 10μM
|
|
Z80548 |
Z80548
|
THP-1 (Acute Monocytic Leukemia Cells) |
1830 |
0.19 |
Functional ≤ 10μM
|
|
Z80559 |
Z80559
|
U251 |
500 |
0.21 |
Functional ≤ 10μM
|
|
Z80600 |
Z80600
|
XF498 (Glioma Cells) |
1720 |
0.19 |
Functional ≤ 10μM
|
|
Z80601 |
Z80601
|
XRS6 |
140 |
0.23 |
Functional ≤ 10μM
|
|
Z80604 |
Z80604
|
YAPC |
1320 |
0.20 |
Functional ≤ 10μM
|
|
Z80608 |
Z80608
|
ZR-75-1 (Breast Carcinoma Cells) |
240 |
0.22 |
Functional ≤ 10μM
|
|
Z80156 |
Z80156
|
HL-60 (Promyeloblast Leukemia Cells) |
300 |
0.22 |
ADME/T ≤ 10μM
|
|
Z80291 |
Z80291
|
MRC5 (Embryonic Lung Fibroblast Cells) |
3900 |
0.18 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.70 |
-7.49 |
-18.52 |
3 |
13 |
0 |
160 |
588.562 |
5 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
4252643
-
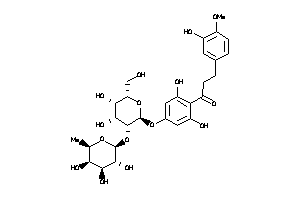
-
4252672
-
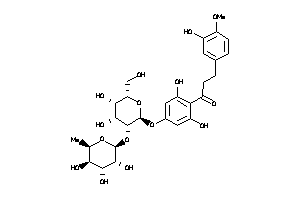
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.01 |
-18.71 |
-23.9 |
9 |
15 |
0 |
245 |
612.581 |
10 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.56 |
2.64 |
-11.61 |
2 |
4 |
0 |
71 |
254.241 |
1 |
↓
|
|
|
|
|
|
|
|
|
|
|