|
|
Analogs
-
35459111
-
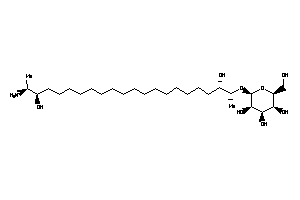
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.44 |
-1.21 |
-5.95 |
4 |
6 |
0 |
99 |
320.426 |
11 |
↓
|
|
|
|
Analogs
-
8637880
-
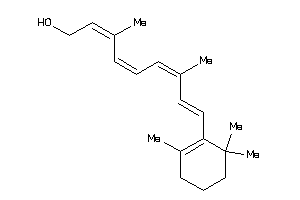
-
12484934
-
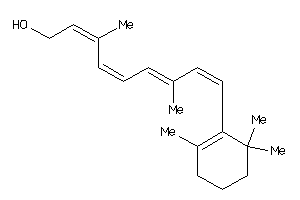
-
12496764
-
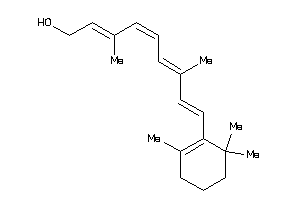
-
12496767
-
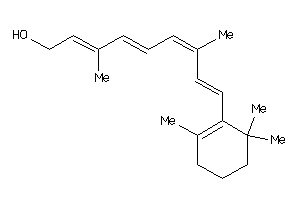
-
12496774
-
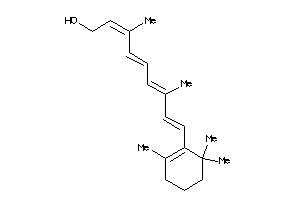
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LACB-1-E |
Beta-lactoglobulin (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
36 |
0.50 |
Binding ≤ 10μM
|
|
RET1-1-E |
Cellular Retinol-binding Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET2-1-E |
Cellular Retinol-binding Protein II (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET3-1-E |
Interstitial Retinol-binding Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET4-1-E |
Plasma Retinol-binding Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET5-1-E |
Cellular Retinol-binding Protein III (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET7-1-E |
Cellular Retinol-binding Protein IV (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
70 |
0.48 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LACB_BOVIN |
P02754
|
Beta-lactoglobulin, Bovin |
36 |
0.50 |
Binding ≤ 1μM
|
|
RET1_HUMAN |
P09455
|
Cellular Retinol-binding Protein, Human |
70 |
0.48 |
Binding ≤ 1μM
|
|
RET2_HUMAN |
P50120
|
Cellular Retinol-binding Protein II, Human |
70 |
0.48 |
Binding ≤ 1μM
|
|
RET5_HUMAN |
P82980
|
Cellular Retinol-binding Protein III, Human |
70 |
0.48 |
Binding ≤ 1μM
|
|
RET7_HUMAN |
Q96R05
|
Cellular Retinol-binding Protein IV, Human |
70 |
0.48 |
Binding ≤ 1μM
|
|
RET3_HUMAN |
P10745
|
Interstitial Retinol-binding Protein, Human |
70 |
0.48 |
Binding ≤ 1μM
|
|
RET4_HUMAN |
P02753
|
Plasma Retinol-binding Protein, Human |
70 |
0.48 |
Binding ≤ 1μM
|
|
LACB_BOVIN |
P02754
|
Beta-lactoglobulin, Bovin |
36 |
0.50 |
Binding ≤ 10μM
|
|
RET1_HUMAN |
P09455
|
Cellular Retinol-binding Protein, Human |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET2_HUMAN |
P50120
|
Cellular Retinol-binding Protein II, Human |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET5_HUMAN |
P82980
|
Cellular Retinol-binding Protein III, Human |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET7_HUMAN |
Q96R05
|
Cellular Retinol-binding Protein IV, Human |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET3_HUMAN |
P10745
|
Interstitial Retinol-binding Protein, Human |
70 |
0.48 |
Binding ≤ 10μM
|
|
RET4_HUMAN |
P02753
|
Plasma Retinol-binding Protein, Human |
70 |
0.48 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.92 |
9.9 |
-4.1 |
1 |
1 |
0 |
20 |
286.459 |
5 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
8218788
-
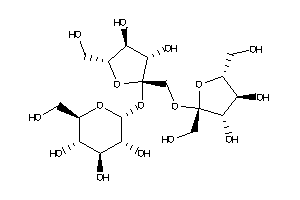
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.90 |
-23.65 |
-19.26 |
11 |
16 |
0 |
269 |
504.438 |
8 |
↓
|
|
|
|
Analogs
-
3872686
-
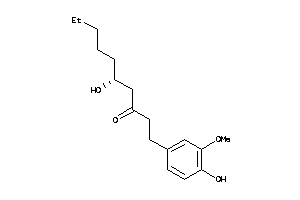
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.22 |
6.04 |
-12.03 |
2 |
4 |
0 |
67 |
294.391 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.95 |
1.62 |
-17.14 |
0 |
6 |
0 |
63 |
364.353 |
3 |
↓
|
|
|
|
Analogs
-
1258940
-
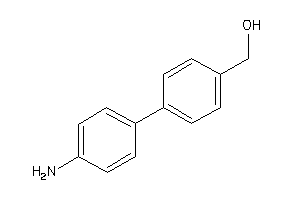
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.35 |
-0.3 |
-5.03 |
3 |
2 |
0 |
46 |
123.155 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R-2-E |
Adenosine A1 Receptor (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
863 |
0.42 |
Binding ≤ 10μM
|
|
AA2AR-3-E |
Adenosine A2a Receptor (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
966 |
0.42 |
Binding ≤ 10μM
|
|
AA2BR-1-E |
Adenosine A2b Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
966 |
0.42 |
Binding ≤ 10μM
|
|
AA3R-4-E |
Adenosine Receptor A3 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
3150 |
0.39 |
Binding ≤ 10μM
|
|
CHLE-1-E |
Butyrylcholinesterase (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
6900 |
0.36 |
Binding ≤ 10μM
|
|
CP1B1-1-E |
Cytochrome P450 1B1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
25 |
0.53 |
Binding ≤ 10μM
|
|
MDR3-1-E |
P-glycoprotein 3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5900 |
0.37 |
Binding ≤ 10μM
|
|
XDH-2-E |
Xanthine Dehydrogenase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1800 |
0.40 |
Binding ≤ 10μM
|
|
CP1A1-1-E |
Cytochrome P450 1A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
77 |
0.50 |
ADME/T ≤ 10μM
|
|
CP1A2-1-E |
Cytochrome P450 1A2 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
40 |
0.52 |
ADME/T ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA1R_RAT |
P25099
|
Adenosine A1 Receptor, Rat |
863 |
0.42 |
Binding ≤ 1μM
|
|
AA2AR_RAT |
P30543
|
Adenosine A2a Receptor, Rat |
966 |
0.42 |
Binding ≤ 1μM
|
|
AA2BR_RAT |
P29276
|
Adenosine A2b Receptor, Rat |
966 |
0.42 |
Binding ≤ 1μM
|
|
CP1B1_HUMAN |
Q16678
|
Cytochrome P450 1B1, Human |
25 |
0.53 |
Binding ≤ 1μM
|
|
AA1R_RAT |
P25099
|
Adenosine A1 Receptor, Rat |
863 |
0.42 |
Binding ≤ 10μM
|
|
AA2AR_RAT |
P30543
|
Adenosine A2a Receptor, Rat |
966 |
0.42 |
Binding ≤ 10μM
|
|
AA2BR_RAT |
P29276
|
Adenosine A2b Receptor, Rat |
966 |
0.42 |
Binding ≤ 10μM
|
|
AA3R_HUMAN |
P33765
|
Adenosine A3 Receptor, Human |
3150 |
0.39 |
Binding ≤ 10μM
|
|
CHLE_HUMAN |
P06276
|
Butyrylcholinesterase, Human |
6900 |
0.36 |
Binding ≤ 10μM
|
|
CP1B1_HUMAN |
Q16678
|
Cytochrome P450 1B1, Human |
25 |
0.53 |
Binding ≤ 10μM
|
|
MDR3_MOUSE |
P21447
|
P-glycoprotein 3, Mouse |
5900 |
0.37 |
Binding ≤ 10μM
|
|
XDH_HUMAN |
P47989
|
Xanthine Dehydrogenase, Human |
1800 |
0.40 |
Binding ≤ 10μM
|
|
CP1A1_HUMAN |
P04798
|
Cytochrome P450 1A1, Human |
77 |
0.50 |
ADME/T ≤ 10μM
|
|
CP1A2_HUMAN |
P05177
|
Cytochrome P450 1A2, Human |
40 |
0.52 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.65 |
2.03 |
-10.35 |
3 |
5 |
0 |
91 |
270.24 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.65 |
2.85 |
-56.02 |
2 |
5 |
-1 |
94 |
269.232 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CCNB1-1-E |
G2/mitotic-specific Cyclin B1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
790 |
0.41 |
Binding ≤ 10μM
|
|
CCNB2-1-E |
G2/mitotic-specific Cyclin B2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
790 |
0.41 |
Binding ≤ 10μM
|
|
CCNB3-1-E |
G2/mitotic-specific Cyclin B3 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
790 |
0.41 |
Binding ≤ 10μM
|
|
CDK1-1-E |
Cyclin-dependent Kinase 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
790 |
0.41 |
Binding ≤ 10μM
|
|
CDK2-1-E |
Cyclin-dependent Kinase 2 (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
5000 |
0.35 |
Binding ≤ 10μM
|
|
CDK6-1-E |
Cyclin-dependent Kinase 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
850 |
0.40 |
Binding ≤ 10μM
|
|
GSK3A-1-E |
Glycogen Synthase Kinase-3 Alpha (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
420 |
0.43 |
Binding ≤ 10μM
|
|
GSK3B-7-E |
Glycogen Synthase Kinase-3 Beta (cluster #7 Of 7), Eukaryotic |
Eukaryotes |
420 |
0.43 |
Binding ≤ 10μM
|
|
LOX12-2-E |
Arachidonate 12-lipoxygenase (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
950 |
0.40 |
Binding ≤ 10μM
|
|
LOX15-1-E |
Arachidonate 15-lipoxygenase (cluster #1 Of 5), Eukaryotic |
Eukaryotes |
1400 |
0.39 |
Binding ≤ 10μM
|
|
Q965D5-1-E |
Enoyl-acyl-carrier Protein Reductase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1000 |
0.40 |
Binding ≤ 10μM
|
|
Q965D6-1-E |
3-oxoacyl-acyl-carrier Protein Reductase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4100 |
0.36 |
Binding ≤ 10μM
|
|
Q965D7-2-E |
Fatty Acid Synthase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2000 |
0.38 |
Binding ≤ 10μM
|
|
XDH-2-E |
Xanthine Dehydrogenase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4330 |
0.36 |
Binding ≤ 10μM
|
|
Z104294-2-O |
Cyclin-dependent Kinase 5/CDK5 Activator 1 (cluster #2 Of 2), Other |
Other |
570 |
0.42 |
Binding ≤ 10μM
|
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
8200 |
0.34 |
Functional ≤ 10μM
|
|
Z50597-1-O |
Rattus Norvegicus (cluster #1 Of 12), Other |
Other |
200 |
0.45 |
Functional ≤ 10μM
|
|
Q7ZJM1-1-V |
Human Immunodeficiency Virus Type 1 Integrase (cluster #1 Of 6), Viral |
Viruses |
8500 |
0.34 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
LOX12_HUMAN |
P18054
|
Arachidonate 12-lipoxygenase, Human |
950 |
0.40 |
Binding ≤ 1μM
|
|
CDK1_HUMAN |
P06493
|
Cyclin-dependent Kinase 1, Human |
790 |
0.41 |
Binding ≤ 1μM
|
|
Z104294 |
Z104294
|
Cyclin-dependent Kinase 5/CDK5 Activator 1 |
570 |
0.42 |
Binding ≤ 1μM
|
|
CDK6_HUMAN |
Q00534
|
Cyclin-dependent Kinase 6, Human |
850 |
0.40 |
Binding ≤ 1μM
|
|
Q965D5_PLAFA |
Q965D5
|
Enoyl-acyl-carrier Protein Reductase, Plafa |
1000 |
0.40 |
Binding ≤ 1μM
|
|
CCNB1_HUMAN |
P14635
|
G2/mitotic-specific Cyclin B1, Human |
790 |
0.41 |
Binding ≤ 1μM
|
|
CCNB2_HUMAN |
O95067
|
G2/mitotic-specific Cyclin B2, Human |
790 |
0.41 |
Binding ≤ 1μM
|
|
CCNB3_HUMAN |
Q8WWL7
|
G2/mitotic-specific Cyclin B3, Human |
790 |
0.41 |
Binding ≤ 1μM
|
|
GSK3A_HUMAN |
P49840
|
Glycogen Synthase Kinase-3 Alpha, Human |
420 |
0.43 |
Binding ≤ 1μM
|
|
GSK3B_HUMAN |
P49841
|
Glycogen Synthase Kinase-3 Beta, Human |
420 |
0.43 |
Binding ≤ 1μM
|
|
Q965D6_PLAFA |
Q965D6
|
3-oxoacyl-acyl-carrier Protein Reductase, Plafa |
4100 |
0.36 |
Binding ≤ 10μM
|
|
LOX12_HUMAN |
P18054
|
Arachidonate 12-lipoxygenase, Human |
950 |
0.40 |
Binding ≤ 10μM
|
|
LOX15_HUMAN |
P16050
|
Arachidonate 15-lipoxygenase, Human |
1400 |
0.39 |
Binding ≤ 10μM
|
|
CDK1_HUMAN |
P06493
|
Cyclin-dependent Kinase 1, Human |
790 |
0.41 |
Binding ≤ 10μM
|
|
CDK2_HUMAN |
P24941
|
Cyclin-dependent Kinase 2, Human |
5000 |
0.35 |
Binding ≤ 10μM
|
|
Z104294 |
Z104294
|
Cyclin-dependent Kinase 5/CDK5 Activator 1 |
570 |
0.42 |
Binding ≤ 10μM
|
|
CDK6_HUMAN |
Q00534
|
Cyclin-dependent Kinase 6, Human |
850 |
0.40 |
Binding ≤ 10μM
|
|
Q965D5_PLAFA |
Q965D5
|
Enoyl-acyl-carrier Protein Reductase, Plafa |
1000 |
0.40 |
Binding ≤ 10μM
|
|
Q965D7_PLAFA |
Q965D7
|
Fatty Acid Synthase, Plafa |
2000 |
0.38 |
Binding ≤ 10μM
|
|
CCNB1_HUMAN |
P14635
|
G2/mitotic-specific Cyclin B1, Human |
790 |
0.41 |
Binding ≤ 10μM
|
|
CCNB2_HUMAN |
O95067
|
G2/mitotic-specific Cyclin B2, Human |
790 |
0.41 |
Binding ≤ 10μM
|
|
CCNB3_HUMAN |
Q8WWL7
|
G2/mitotic-specific Cyclin B3, Human |
790 |
0.41 |
Binding ≤ 10μM
|
|
GSK3A_HUMAN |
P49840
|
Glycogen Synthase Kinase-3 Alpha, Human |
420 |
0.43 |
Binding ≤ 10μM
|
|
GSK3B_HUMAN |
P49841
|
Glycogen Synthase Kinase-3 Beta, Human |
420 |
0.43 |
Binding ≤ 10μM
|
|
Q7ZJM1_9HIV1 |
Q7ZJM1
|
Human Immunodeficiency Virus Type 1 Integrase, 9hiv1 |
8500 |
0.34 |
Binding ≤ 10μM
|
|
XDH_HUMAN |
P47989
|
Xanthine Dehydrogenase, Human |
4330 |
0.36 |
Binding ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
6500 |
0.35 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
200 |
0.45 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.97 |
-1.09 |
-11.69 |
4 |
6 |
0 |
111 |
286.239 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.97 |
-0.3 |
-54.07 |
3 |
6 |
-1 |
114 |
285.231 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.07 |
6.73 |
-2.9 |
1 |
1 |
0 |
20 |
220.356 |
4 |
↓
|
|
|
|
Analogs
-
17002804
-
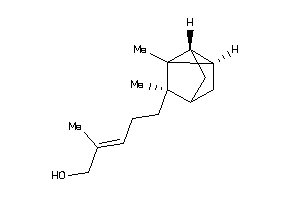
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.07 |
6.72 |
-2.91 |
1 |
1 |
0 |
20 |
220.356 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.70 |
-18.02 |
-14.27 |
9 |
11 |
0 |
201 |
344.313 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.30 |
8.89 |
-5.28 |
0 |
0 |
0 |
0 |
178.234 |
0 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81247-1-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #1 Of 9), Other |
Other |
7800 |
0.31 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z81247 |
Z81247
|
HeLa (Cervical Adenocarcinoma Cells) |
7800 |
0.31 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.22 |
-0.74 |
-12.4 |
4 |
7 |
0 |
120 |
316.265 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.22 |
0.25 |
-52.59 |
3 |
7 |
-1 |
123 |
315.257 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.22 |
-0.4 |
-54.62 |
3 |
7 |
-1 |
123 |
315.257 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.79 |
-16.25 |
-19.66 |
8 |
11 |
0 |
197 |
342.297 |
8 |
↓
|
|
Ref
Reference (pH 7)
|
-4.45 |
-17.15 |
-12.45 |
8 |
11 |
0 |
197 |
342.297 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
-4.79 |
-15.49 |
-66.22 |
7 |
11 |
-1 |
200 |
341.289 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80390-1-O |
PC-3 (Prostate Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
7200 |
0.26 |
ADME/T ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80390 |
Z80390
|
PC-3 (Prostate Carcinoma Cells) |
7200 |
0.26 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.66 |
5.65 |
-15.74 |
1 |
7 |
0 |
83 |
380.352 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.66 |
6.41 |
-41.24 |
0 |
7 |
-1 |
86 |
379.344 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.47 |
-2.58 |
-30.81 |
8 |
5 |
1 |
104 |
246.423 |
12 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-5.47 |
-2.2 |
-103.97 |
9 |
5 |
2 |
106 |
247.431 |
12 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-5.47 |
-1.81 |
-183.15 |
10 |
5 |
3 |
107 |
248.439 |
12 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.23 |
-3.8 |
-11.84 |
3 |
3 |
0 |
63 |
89.094 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.23 |
-3.85 |
-8.49 |
3 |
3 |
0 |
63 |
89.094 |
1 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.82 |
4.67 |
-4.91 |
0 |
2 |
0 |
26 |
116.16 |
4 |
↓
|
|
|
|
Analogs
-
32840814
-
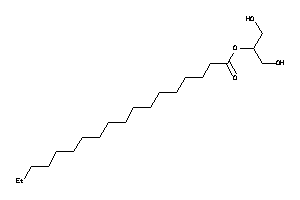
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.09 |
-0.33 |
-7.92 |
2 |
4 |
0 |
66 |
330.509 |
18 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
10.67 |
38.95 |
-9.43 |
0 |
6 |
0 |
79 |
807.339 |
50 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.30 |
-14.62 |
-15.27 |
8 |
11 |
0 |
197 |
342.297 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.57 |
-18.99 |
-37.57 |
11 |
19 |
0 |
295 |
627.597 |
10 |
↓
|
|
Ref
Reference (pH 7)
|
-5.18 |
-30.58 |
-31.69 |
11 |
19 |
0 |
306 |
627.597 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.01 |
8.6 |
-0.53 |
0 |
0 |
0 |
0 |
204.357 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.27 |
-3.35 |
-15.46 |
1 |
2 |
0 |
37 |
88.106 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.58 |
0.13 |
-3.03 |
1 |
1 |
0 |
20 |
72.107 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.56 |
5.76 |
-5.02 |
0 |
1 |
0 |
17 |
126.199 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
2.56 |
5.73 |
-4.35 |
0 |
1 |
0 |
17 |
126.199 |
4 |
↓
|
|
|
|
|
|
|
|
|
|
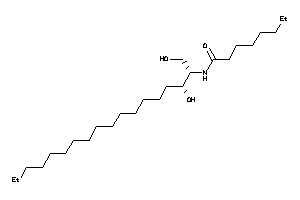
Analogs
-
43552983
-
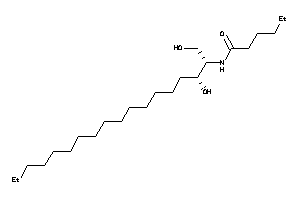
-
8860496
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
9.81 |
17.74 |
-7.11 |
3 |
4 |
0 |
70 |
539.93 |
31 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.33 |
2.22 |
-11.05 |
0 |
3 |
0 |
51 |
300.398 |
1 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.77 |
3.84 |
-4.03 |
0 |
2 |
0 |
34 |
176.986 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.34 |
2.29 |
-5.98 |
0 |
1 |
0 |
17 |
126.199 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.61 |
13.77 |
-6.43 |
0 |
3 |
0 |
36 |
391.294 |
7 |
↓
|
|
|
|
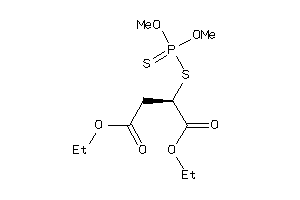
Analogs
-
1530800
-
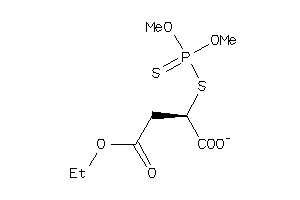
-
32153077
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.32 |
11.04 |
-17.77 |
0 |
6 |
0 |
71 |
330.364 |
11 |
↓
|
|