|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.30 |
13.09 |
-1.44 |
1 |
1 |
0 |
20 |
400.691 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.75 |
7.92 |
-47.26 |
1 |
3 |
-1 |
60 |
317.449 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.75 |
5.92 |
-4.29 |
2 |
3 |
0 |
58 |
318.457 |
1 |
↓
|
|
|
|
Analogs
-
1698429
-
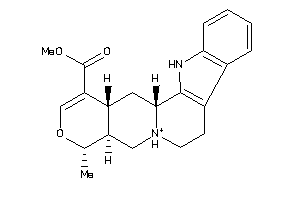
-
3978730
-
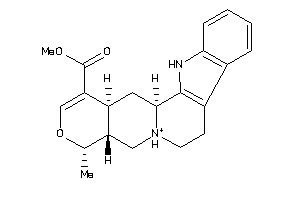
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ADA2A-1-E |
Alpha-2a Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8 |
0.44 |
Binding ≤ 10μM
|
|
ADA2B-1-E |
Alpha-2b Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
15 |
0.42 |
Binding ≤ 10μM
|
|
ADA2C-1-E |
Alpha-2c Adrenergic Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
5 |
0.45 |
Binding ≤ 10μM
|
|
CAC1C-1-E |
Voltage-gated L-type Calcium Channel Alpha-1C Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
289 |
0.35 |
Binding ≤ 10μM
|
|
CP2D6-1-E |
Cytochrome P450 2D6 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6 |
0.44 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Popular Name:
(3S,4aR,6aR,6bS,8aR,11R,12S,12aR,14aS,14bR)-4,4,6a,6b,8a,11,12,14b-octamethyl-2,3,4a,5,6,7,8,9,10,11
(3S,4aR,6aR,6bS,8aR,11R,12S,12aR…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
8.08 |
12.44 |
-1.17 |
1 |
1 |
0 |
20 |
426.729 |
0 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.41 |
5.3 |
-12.9 |
0 |
6 |
0 |
85 |
228.229 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.21 |
2.23 |
-13.07 |
0 |
8 |
0 |
111 |
332.359 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.00 |
-1.66 |
-43.22 |
0 |
4 |
-1 |
65 |
203.63 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.79 |
5.27 |
-51.98 |
0 |
6 |
-1 |
99 |
213.194 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.81 |
6.68 |
-8.94 |
0 |
6 |
0 |
85 |
276.701 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.78 |
6.27 |
-12.37 |
0 |
6 |
0 |
85 |
242.256 |
6 |
↓
|
|
|
|
Analogs
-
4632590
-
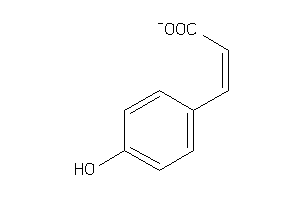
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
1070 |
0.70 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
8010 |
0.59 |
Binding ≤ 10μM
|
|
CAH14-4-E |
Carbonic Anhydrase XIV (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
6680 |
0.60 |
Binding ≤ 10μM
|
|
CAH2-13-E |
Carbonic Anhydrase II (cluster #13 Of 15), Eukaryotic |
Eukaryotes |
980 |
0.70 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
7570 |
0.60 |
Binding ≤ 10μM
|
|
CAH4-3-E |
Carbonic Anhydrase IV (cluster #3 Of 16), Eukaryotic |
Eukaryotes |
9600 |
0.59 |
Binding ≤ 10μM
|
|
CAH5A-8-E |
Carbonic Anhydrase VA (cluster #8 Of 10), Eukaryotic |
Eukaryotes |
5960 |
0.61 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
7760 |
0.60 |
Binding ≤ 10μM
|
|
CAH6-2-E |
Carbonic Anhydrase VI (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
6720 |
0.60 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
5230 |
0.62 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
5330 |
0.62 |
Binding ≤ 10μM
|
|
Z50594-8-O |
Mus Musculus (cluster #8 Of 9), Other |
Other |
190 |
0.78 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
980 |
0.70 |
Binding ≤ 1μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
1070 |
0.70 |
Binding ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
980 |
0.70 |
Binding ≤ 10μM
|
|
CAH3_HUMAN |
P07451
|
Carbonic Anhydrase III, Human |
7570 |
0.60 |
Binding ≤ 10μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
9600 |
0.59 |
Binding ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
5330 |
0.62 |
Binding ≤ 10μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
5960 |
0.61 |
Binding ≤ 10μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
7760 |
0.60 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
6720 |
0.60 |
Binding ≤ 10μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
5230 |
0.62 |
Binding ≤ 10μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
8010 |
0.59 |
Binding ≤ 10μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
6680 |
0.60 |
Binding ≤ 10μM
|
|
Z50594 |
Z50594
|
Mus Musculus |
190 |
0.78 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.43 |
2.5 |
-50.43 |
1 |
3 |
-1 |
60 |
163.152 |
2 |
↓
|
|
|
|
Analogs
-
39289888
-
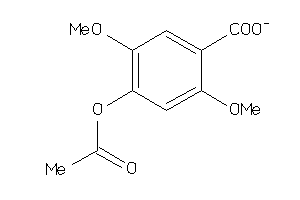
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.04 |
5.67 |
-64.52 |
0 |
8 |
-1 |
119 |
295.223 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.43 |
5.36 |
-56.15 |
0 |
4 |
-1 |
66 |
497.74 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.50 |
3.65 |
-26.73 |
1 |
5 |
1 |
55 |
260.269 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.50 |
2.62 |
-11.37 |
0 |
5 |
0 |
54 |
259.261 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.85 |
2.75 |
-29.23 |
5 |
3 |
1 |
64 |
128.199 |
3 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.53 |
10.44 |
-51.21 |
0 |
3 |
-1 |
49 |
287.379 |
7 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.51 |
5.75 |
-45.52 |
0 |
5 |
-1 |
68 |
239.247 |
6 |
↓
|
|
|
|
Analogs
-
8829465
-
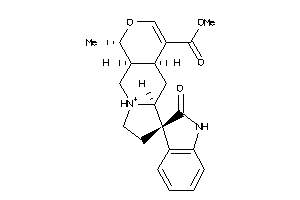
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.18 |
0.99 |
-49.14 |
2 |
6 |
1 |
69 |
369.441 |
2 |
↓
|
|
|
|
Analogs
-
8829465
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.18 |
7.93 |
-48.95 |
2 |
6 |
1 |
69 |
369.441 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.18 |
5.66 |
-9.91 |
1 |
6 |
0 |
68 |
368.433 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.54 |
5.5 |
-47.59 |
0 |
5 |
-1 |
68 |
237.231 |
5 |
↓
|
|
|
|
Analogs
-
12410298
-
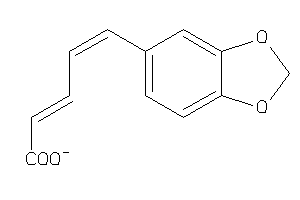
-
13099346
-
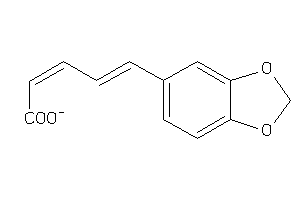
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.32 |
0.57 |
-52.07 |
0 |
4 |
-1 |
59 |
217.2 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ALDR-2-E |
Aldose Reductase (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
3500 |
0.69 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.30 |
2.13 |
-51.49 |
1 |
3 |
-1 |
60 |
151.141 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
9920 |
0.70 |
Binding ≤ 10μM
|
|
CAH12-9-E |
Carbonic Anhydrase XII (cluster #9 Of 9), Eukaryotic |
Eukaryotes |
8800 |
0.71 |
Binding ≤ 10μM
|
|
CAH15-4-E |
Carbonic Anhydrase 15 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
7200 |
0.72 |
Binding ≤ 10μM
|
|
CAH2-13-E |
Carbonic Anhydrase II (cluster #13 Of 15), Eukaryotic |
Eukaryotes |
7120 |
0.72 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.87 |
3.11 |
-51.26 |
1 |
3 |
-1 |
60 |
137.114 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.15 |
-12.92 |
-54.49 |
5 |
10 |
-1 |
168 |
373.334 |
5 |
↓
|
|
|
|
Analogs
-
33831555
-
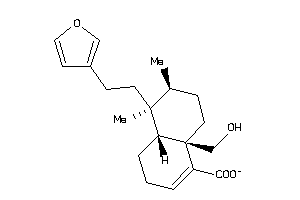
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.67 |
10.86 |
-53.01 |
0 |
3 |
-1 |
53 |
315.433 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
4.67 |
9.03 |
-7.28 |
1 |
3 |
0 |
50 |
316.441 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.19 |
2.2 |
-46.94 |
1 |
4 |
-1 |
70 |
167.14 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
1080 |
0.76 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
4090 |
0.69 |
Binding ≤ 10μM
|
|
CAH2-13-E |
Carbonic Anhydrase II (cluster #13 Of 15), Eukaryotic |
Eukaryotes |
470 |
0.81 |
Binding ≤ 10μM
|
|
CAH6-8-E |
Carbonic Anhydrase VI (cluster #8 Of 8), Eukaryotic |
Eukaryotes |
4720 |
0.68 |
Binding ≤ 10μM |
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
4450 |
0.68 |
Binding ≤ 10μM
|
|
Q2PCB5-2-E |
Carbonic Anhydrase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3210 |
0.70 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.88 |
0.14 |
-48.46 |
2 |
4 |
-1 |
81 |
153.113 |
1 |
↓
|
|
|
|
Analogs
-
3947453
-
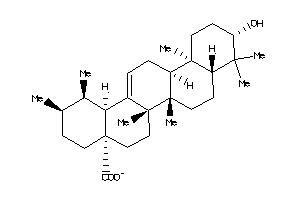
-
3947454
-
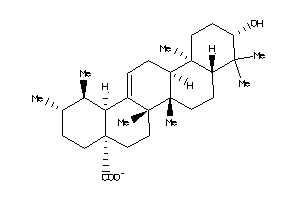
-
3947455
-
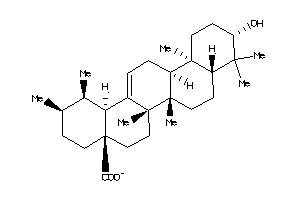
-
3947456
-
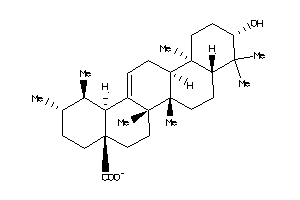
-
3978825
-
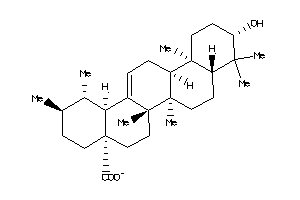
Draw
Identity
99%
90%
80%
70%
Popular Name:
10-hydroxy-1,2,6a,6b,9,9,12a-heptamethyl-2,3,4,5,6,6a,7,8,8a,10,11,12,13,14b-tetradecahydro-1H-picen
10-hydroxy-1,2,6a,6b,9,9,12a-hep…
Find On:
PubMed —
Wikipedia —
Google
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.79 |
1.9 |
-46.86 |
1 |
3 |
-1 |
60 |
455.703 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.05 |
5.92 |
-50.73 |
0 |
3 |
-1 |
49 |
203.217 |
1 |
↓
|
|
|
|
Analogs
-
4716552
-
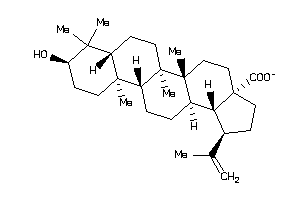
-
4995154
-
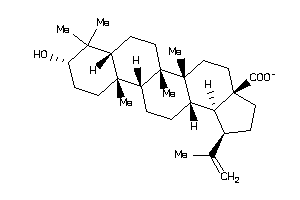
-
4995155
-
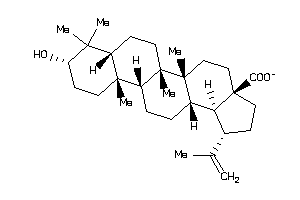
-
4995156
-
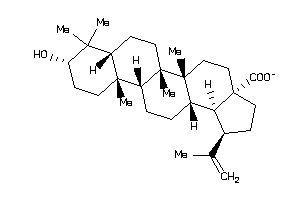
-
8951991
-
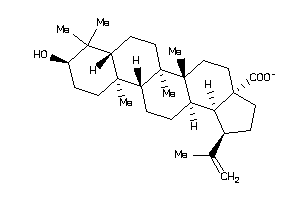
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DPOLB-2-E |
DNA Polymerase Beta (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
6500 |
0.22 |
Binding ≤ 10μM
|
|
GPBAR-2-E |
G-protein Coupled Bile Acid Receptor 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2170 |
0.24 |
Functional ≤ 10μM
|
|
NR1H4-2-E |
Bile Acid Receptor FXR (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
0 |
0.00 |
Functional ≤ 10μM
|
|
Z103202-1-O |
8505C (cluster #1 Of 2), Other |
Other |
7260 |
0.22 |
Functional ≤ 10μM
|
|
Z50425-4-O |
Plasmodium Falciparum (cluster #4 Of 22), Other |
Other |
5200 |
0.22 |
Functional ≤ 10μM
|
|
Z50459-6-O |
Leishmania Donovani (cluster #6 Of 8), Other |
Other |
4100 |
0.23 |
Functional ≤ 10μM
|
|
Z50602-3-O |
Human Herpesvirus 1 (cluster #3 Of 5), Other |
Other |
8200 |
0.22 |
Functional ≤ 10μM
|
|
Z50607-8-O |
Human Immunodeficiency Virus 1 (cluster #8 Of 10), Other |
Other |
3100 |
0.23 |
Functional ≤ 10μM
|
|
Z50636-1-O |
Sindbis Virus (cluster #1 Of 3), Other |
Other |
500 |
0.27 |
Functional ≤ 10μM
|
|
Z80482-1-O |
SK-MEL-2 (Melanoma Cells) (cluster #1 Of 4), Other |
Other |
7000 |
0.22 |
Functional ≤ 10μM
|
|
Z80526-1-O |
SW480 (Colon Adenocarcinoma Cells) (cluster #1 Of 6), Other |
Other |
6480 |
0.22 |
Functional ≤ 10μM
|
|
Z80682-3-O |
A549 (Lung Carcinoma Cells) (cluster #3 Of 11), Other |
Other |
6650 |
0.22 |
Functional ≤ 10μM
|
|
Z80897-2-O |
H9 (T-lymphoid Cells) (cluster #2 Of 2), Other |
Other |
1400 |
0.25 |
Functional ≤ 10μM
|
|
Z81020-2-O |
HepG2 (Hepatoblastoma Cells) (cluster #2 Of 8), Other |
Other |
36 |
0.32 |
Functional ≤ 10μM
|
|
Z81072-2-O |
Jurkat (Acute Leukemic T-cells) (cluster #2 Of 10), Other |
Other |
27 |
0.32 |
Functional ≤ 10μM
|
|
Z81247-4-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #4 Of 9), Other |
Other |
26 |
0.32 |
Functional ≤ 10μM
|
|
Z80897-2-O |
H9 (T-lymphoid Cells) (cluster #2 Of 2), Other |
Other |
900 |
0.26 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.04 |
12.1 |
-48.88 |
1 |
3 |
-1 |
60 |
455.703 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
7.04 |
10.34 |
-5.48 |
2 |
3 |
0 |
58 |
456.711 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
3200 |
0.64 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
7780 |
0.60 |
Binding ≤ 10μM
|
|
CAH13-1-E |
Carbonic Anhydrase XIII (cluster #1 Of 7), Eukaryotic |
Eukaryotes |
9860 |
0.58 |
Binding ≤ 10μM
|
|
CAH14-4-E |
Carbonic Anhydrase XIV (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
7030 |
0.60 |
Binding ≤ 10μM
|
|
CAH2-13-E |
Carbonic Anhydrase II (cluster #13 Of 15), Eukaryotic |
Eukaryotes |
2250 |
0.66 |
Binding ≤ 10μM
|
|
CAH3-2-E |
Carbonic Anhydrase III (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
7490 |
0.60 |
Binding ≤ 10μM
|
|
CAH4-3-E |
Carbonic Anhydrase IV (cluster #3 Of 16), Eukaryotic |
Eukaryotes |
9800 |
0.58 |
Binding ≤ 10μM
|
|
CAH5A-6-E |
Carbonic Anhydrase VA (cluster #6 Of 10), Eukaryotic |
Eukaryotes |
4080 |
0.63 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
9970 |
0.58 |
Binding ≤ 10μM
|
|
CAH6-2-E |
Carbonic Anhydrase VI (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
6130 |
0.61 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
6070 |
0.61 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
6990 |
0.60 |
Binding ≤ 10μM
|
|
Q2PCB5-2-E |
Carbonic Anhydrase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4130 |
0.63 |
Binding ≤ 10μM |
|
Z100766-1-O |
Radical Scavenging Activity (cluster #1 Of 2), Other |
Other |
7300 |
0.60 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Q2PCB5_DICLA |
Q2PCB5
|
Carbonic Anhydrase, Dicla |
4130 |
0.63 |
Binding ≤ 10μM |
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
3200 |
0.64 |
Binding ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
2250 |
0.66 |
Binding ≤ 10μM
|
|
CAH3_HUMAN |
P07451
|
Carbonic Anhydrase III, Human |
7490 |
0.60 |
Binding ≤ 10μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
9800 |
0.58 |
Binding ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
6990 |
0.60 |
Binding ≤ 10μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
4080 |
0.63 |
Binding ≤ 10μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
9970 |
0.58 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
6130 |
0.61 |
Binding ≤ 10μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
6070 |
0.61 |
Binding ≤ 10μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
7780 |
0.60 |
Binding ≤ 10μM
|
|
CAH13_MOUSE |
Q9D6N1
|
Carbonic Anhydrase XIII, Mouse |
9860 |
0.58 |
Binding ≤ 10μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
7030 |
0.60 |
Binding ≤ 10μM
|
|
Z100766 |
Z100766
|
Radical Scavenging Activity |
6000 |
0.61 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.59 |
-2.02 |
-47.55 |
3 |
5 |
-1 |
101 |
169.112 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.45 |
-1.57 |
-53.75 |
5 |
9 |
-1 |
168 |
353.303 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z102317-1-O |
Brain (cluster #1 Of 2), Other |
Other |
4770 |
0.47 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.26 |
2.4 |
-49.65 |
1 |
5 |
-1 |
79 |
223.204 |
4 |
↓
|
|
|
|
Analogs
-
44201201
-
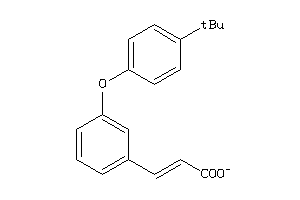
-
44201202
-
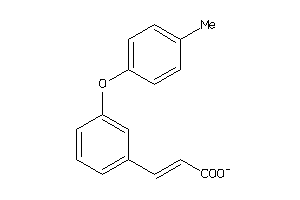
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.25 |
3.5 |
-48.23 |
1 |
4 |
-1 |
70 |
193.178 |
3 |
↓
|
|
|
|
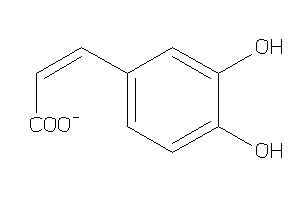
Analogs
-
4482687
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-4-E |
Carbonic Anhydrase I (cluster #4 Of 12), Eukaryotic |
Eukaryotes |
2380 |
0.61 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
9060 |
0.54 |
Binding ≤ 10μM
|
|
CAH14-4-E |
Carbonic Anhydrase XIV (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
8710 |
0.54 |
Binding ≤ 10μM
|
|
CAH2-13-E |
Carbonic Anhydrase II (cluster #13 Of 15), Eukaryotic |
Eukaryotes |
1610 |
0.62 |
Binding ≤ 10μM
|
|
CAH3-1-E |
Carbonic Anhydrase III (cluster #1 Of 6), Eukaryotic |
Eukaryotes |
10000 |
0.54 |
Binding ≤ 10μM
|
|
CAH5A-6-E |
Carbonic Anhydrase VA (cluster #6 Of 10), Eukaryotic |
Eukaryotes |
6490 |
0.56 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
9080 |
0.54 |
Binding ≤ 10μM
|
|
CAH6-2-E |
Carbonic Anhydrase VI (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
7330 |
0.55 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
6420 |
0.56 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
7870 |
0.55 |
Binding ≤ 10μM
|
|
LOX1-1-E |
Seed Lipoxygenase-1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
3000 |
0.59 |
Binding ≤ 10μM
|
|
O49150-1-E |
5-lipoxygenase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
4000 |
0.58 |
Binding ≤ 10μM
|
|
PTN1-1-E |
Protein-tyrosine Phosphatase 1B (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
3060 |
0.59 |
Binding ≤ 10μM
|
|
Z100766-1-O |
Radical Scavenging Activity (cluster #1 Of 2), Other |
Other |
2900 |
0.60 |
Functional ≤ 10μM
|
|
Z50185-2-O |
Staphylococcus Aureus (cluster #2 Of 4), Other |
Other |
2780 |
0.60 |
Functional ≤ 10μM
|
|
Z50186-1-O |
Staphylococcus Epidermidis (cluster #1 Of 2), Other |
Other |
2780 |
0.60 |
Functional ≤ 10μM
|
|
Z50594-8-O |
Mus Musculus (cluster #8 Of 9), Other |
Other |
190 |
0.72 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
O49150_SOLTU |
O49150
|
5-lipoxygenase, Soltu |
3000 |
0.59 |
Binding ≤ 10μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
2380 |
0.61 |
Binding ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
1610 |
0.62 |
Binding ≤ 10μM
|
|
CAH3_HUMAN |
P07451
|
Carbonic Anhydrase III, Human |
10000 |
0.54 |
Binding ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
7870 |
0.55 |
Binding ≤ 10μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
6490 |
0.56 |
Binding ≤ 10μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
9080 |
0.54 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
7330 |
0.55 |
Binding ≤ 10μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
6420 |
0.56 |
Binding ≤ 10μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
9060 |
0.54 |
Binding ≤ 10μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
8710 |
0.54 |
Binding ≤ 10μM
|
|
PTN1_HUMAN |
P18031
|
Protein-tyrosine Phosphatase 1B, Human |
3060 |
0.59 |
Binding ≤ 10μM
|
|
LOX1_SOYBN |
P08170
|
Seed Lipoxygenase-1, Soybn |
3000 |
0.59 |
Binding ≤ 10μM
|
|
Z50594 |
Z50594
|
Mus Musculus |
190 |
0.72 |
Functional ≤ 10μM
|
|
Z100766 |
Z100766
|
Radical Scavenging Activity |
2800 |
0.60 |
Functional ≤ 10μM
|
|
Z50185 |
Z50185
|
Staphylococcus Aureus |
2780 |
0.60 |
Functional ≤ 10μM
|
|
Z50186 |
Z50186
|
Staphylococcus Epidermidis |
2780 |
0.60 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.94 |
1.43 |
-49.59 |
2 |
4 |
-1 |
81 |
179.151 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
6.73 |
12.27 |
-52.74 |
1 |
3 |
-1 |
60 |
455.703 |
1 |
↓
|
|
Lo
Low (pH 4.5-6)
|
6.73 |
10.3 |
-5.37 |
2 |
3 |
0 |
58 |
456.711 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.54 |
-2.68 |
-51.94 |
4 |
13 |
-1 |
201 |
463.412 |
10 |
↓
|
|
|
|
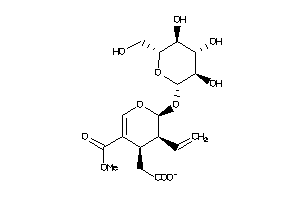
Analogs
-
44681098
-
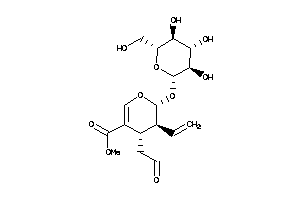
-
16957580
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.33 |
-4.46 |
-62.02 |
4 |
11 |
-1 |
175 |
403.36 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.62 |
6.47 |
-32.22 |
2 |
4 |
0 |
70 |
226.235 |
1 |
↓
|
|
|
|
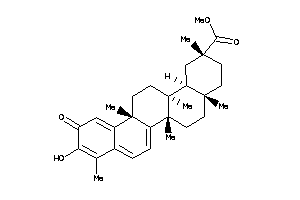
Analogs
-
9212394
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80156-2-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #2 Of 12), Other |
Other |
200 |
0.28 |
Functional ≤ 10μM
|
|
Z80224-2-O |
MCF7 (Breast Carcinoma Cells) (cluster #2 Of 14), Other |
Other |
400 |
0.26 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.67 |
12.78 |
-16.52 |
1 |
4 |
0 |
64 |
464.646 |
2 |
↓
|
|
Ref
Reference (pH 7)
|
6.18 |
10.75 |
-10.5 |
2 |
4 |
0 |
67 |
464.646 |
2 |
↓
|
|
|
|
|
|
|
Analogs
-
39091
-
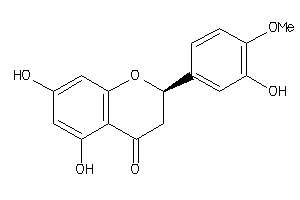
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.16 |
-3.48 |
-11.56 |
3 |
6 |
0 |
96 |
302.282 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TBB1-1-E |
Tubulin Beta-1 Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3000 |
0.32 |
Binding ≤ 10μM
|
|
TBB3-1-E |
Tubulin Beta-3 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.32 |
Binding ≤ 10μM
|
|
TBB4A-1-E |
Tubulin Beta-4 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.32 |
Binding ≤ 10μM
|
|
TBB4B-1-E |
Tubulin Beta-2 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.32 |
Binding ≤ 10μM
|
|
TBB8-1-E |
Tubulin Beta-8 Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
3000 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.27 |
1.16 |
-15.04 |
3 |
7 |
0 |
109 |
330.292 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.53 |
1.45 |
-45.8 |
2 |
7 |
-1 |
112 |
329.284 |
3 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.12 |
-0.02 |
-5.55 |
1 |
3 |
0 |
46 |
190.655 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.89 |
6.11 |
-6.65 |
0 |
3 |
0 |
39 |
231.704 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.99 |
4.25 |
-5.43 |
0 |
4 |
0 |
52 |
232.692 |
5 |
↓
|
|