|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.83 |
2.94 |
-89.88 |
3 |
3 |
0 |
68 |
165.192 |
3 |
↓
|
|
|
|
Analogs
-
39133590
-
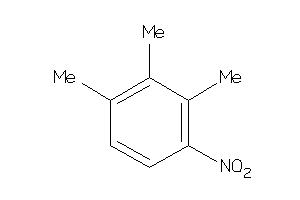
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.10 |
2.56 |
-6.67 |
0 |
3 |
0 |
45 |
165.192 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.08 |
0.8 |
-54.54 |
0 |
3 |
-1 |
43 |
164.184 |
2 |
↓
|
|
|
|
Analogs
-
32202597
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.41 |
2.52 |
-5.75 |
0 |
3 |
0 |
45 |
165.192 |
2 |
↓
|
|
|
|
Analogs
-
39133547
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.10 |
2.79 |
-6.58 |
0 |
3 |
0 |
45 |
165.192 |
1 |
↓
|
|
|
|
|
|
|
Analogs
-
8461952
-
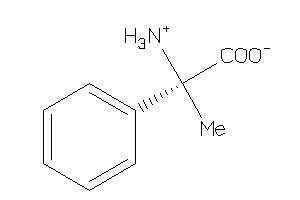
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CA2D1-1-E |
Voltage-gated Calcium Channel Alpha2/delta Subunit 1 (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
170 |
0.79 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.00 |
2.75 |
-38.36 |
3 |
3 |
0 |
68 |
165.192 |
2 |
↓
|
|
|
|
Analogs
-
33957324
-
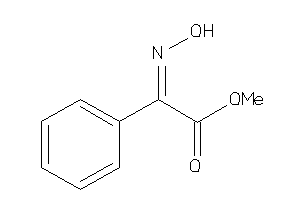
-
5742744
-
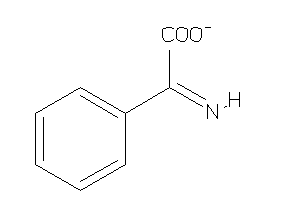
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.77 |
2.71 |
-46.84 |
1 |
4 |
-1 |
73 |
164.14 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.67 |
0.9 |
-50.09 |
2 |
4 |
-1 |
83 |
164.14 |
2 |
↓
|
|
|
|
Analogs
-
4787000
-
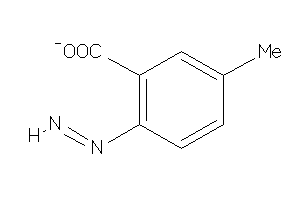
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.21 |
2.04 |
-44.96 |
2 |
4 |
-1 |
83 |
164.14 |
2 |
↓
|
|
|
|
Analogs
-
8617556
-
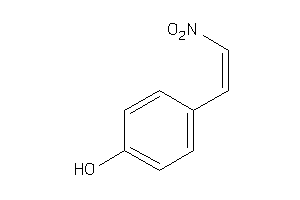
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR-2-E |
Epidermal Growth Factor Receptor ErbB1 (cluster #2 Of 4), Eukaryotic |
Eukaryotes |
10000 |
0.58 |
Binding ≤ 10μM |
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
EGFR_HUMAN |
P00533
|
Epidermal Growth Factor Receptor ErbB1, Human |
10000 |
0.58 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.63 |
3.72 |
-8 |
1 |
4 |
0 |
66 |
165.148 |
2 |
↓
|
|
|
|
Analogs
-
5167589
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.80 |
5.62 |
-9.23 |
0 |
4 |
0 |
63 |
165.148 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.36 |
5.74 |
-45.49 |
0 |
4 |
-1 |
69 |
164.14 |
2 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.24 |
6.31 |
-12.63 |
0 |
4 |
0 |
63 |
165.148 |
3 |
↓
|
|
|
|
Analogs
-
36721159
-

-
36721160
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.77 |
5.68 |
-14.28 |
0 |
4 |
0 |
63 |
165.148 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.28 |
1.84 |
-69.48 |
2 |
4 |
-1 |
83 |
164.14 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.31 |
2.95 |
-48.53 |
0 |
4 |
-1 |
70 |
164.14 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.98 |
6.34 |
-5.07 |
0 |
2 |
0 |
26 |
234.249 |
1 |
↓
|
|
|
|
Analogs
-
4829015
-
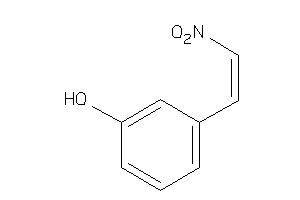
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.60 |
0.43 |
-7.84 |
1 |
4 |
0 |
66 |
165.148 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.87 |
5.71 |
-13.55 |
0 |
4 |
0 |
63 |
165.148 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
MURI-2-B |
Glutamate Racemase (cluster #2 Of 2), Bacterial |
Bacteria |
5800 |
0.73 |
Binding ≤ 10μM
|
|
GRIA1-3-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
1362 |
0.82 |
Binding ≤ 10μM
|
|
GRIA2-3-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
940 |
0.84 |
Binding ≤ 10μM
|
|
GRIA3-3-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
500 |
0.88 |
Binding ≤ 10μM
|
|
GRIA4-3-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #3 Of 5), Eukaryotic |
Eukaryotes |
868 |
0.85 |
Binding ≤ 10μM
|
|
GRIK1-3-E |
Glutamate Receptor Ionotropic Kainate 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
701 |
0.86 |
Binding ≤ 10μM
|
|
GRIK2-2-E |
Glutamate Receptor Ionotropic Kainate 2 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
1106 |
0.83 |
Binding ≤ 10μM
|
|
GRIK3-2-E |
Glutamate Receptor Ionotropic Kainate 3 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
789 |
0.85 |
Binding ≤ 10μM
|
|
GRIK4-2-E |
Glutamate Receptor Ionotropic Kainate 4 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.90 |
Binding ≤ 10μM
|
|
GRIK5-2-E |
Glutamate Receptor Ionotropic Kainate 5 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
400 |
0.90 |
Binding ≤ 10μM
|
|
GRM1-3-E |
Metabotropic Glutamate Receptor 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
4900 |
0.74 |
Binding ≤ 10μM
|
|
GRM2-1-E |
Metabotropic Glutamate Receptor 2 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
290 |
0.92 |
Binding ≤ 10μM
|
|
GRM3-1-E |
Metabotropic Glutamate Receptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
1900 |
0.80 |
Binding ≤ 10μM
|
|
GRM4-3-E |
Metabotropic Glutamate Receptor 4 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
9800 |
0.70 |
Binding ≤ 10μM
|
|
GRM5-4-E |
Metabotropic Glutamate Receptor 5 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
3100 |
0.77 |
Binding ≤ 10μM
|
|
GRM6-1-E |
Metabotropic Glutamate Receptor 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4900 |
0.74 |
Binding ≤ 10μM
|
|
NMDE1-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
200 |
0.94 |
Binding ≤ 10μM
|
|
NMDE2-4-E |
Glutamate [NMDA] Receptor Subunit Epsilon 2 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
200 |
0.94 |
Binding ≤ 10μM
|
|
NMDE3-3-E |
Glutamate [NMDA] Receptor Subunit Epsilon 3 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
200 |
0.94 |
Binding ≤ 10μM
|
|
NMDE4-2-E |
Glutamate [NMDA] Receptor Subunit Epsilon 4 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
200 |
0.94 |
Binding ≤ 10μM
|
|
GRIA1-3-E |
Glutamate Receptor Ionotropic, AMPA 1 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
214 |
0.93 |
Functional ≤ 10μM
|
|
GRIA2-3-E |
Glutamate Receptor Ionotropic, AMPA 2 (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
214 |
0.93 |
Functional ≤ 10μM
|
|
GRIA3-2-E |
Glutamate Receptor Ionotropic, AMPA 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
214 |
0.93 |
Functional ≤ 10μM
|
|
GRIA4-2-E |
Glutamate Receptor Ionotropic, AMPA 4 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
214 |
0.93 |
Functional ≤ 10μM
|
|
GRM1-4-E |
Metabotropic Glutamate Receptor 1 (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
1050 |
0.84 |
Functional ≤ 10μM
|
|
GRM8-1-E |
Metabotropic Glutamate Receptor 8 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
8000 |
0.71 |
Functional ≤ 10μM
|
|
Z104302-3-O |
Glutamate NMDA Receptor (cluster #3 Of 7), Other |
Other |
80 |
0.99 |
Binding ≤ 10μM |
|
Z50597-7-O |
Rattus Norvegicus (cluster #7 Of 12), Other |
Other |
2500 |
0.78 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z104302 |
Z104302
|
Glutamate NMDA Receptor |
70 |
1.00 |
Binding ≤ 1μM
|
|
GRIK1_RAT |
P22756
|
Glutamate Receptor Ionotropic Kainate 1, Rat |
120 |
0.97 |
Binding ≤ 1μM
|
|
GRIK1_HUMAN |
P39086
|
Glutamate Receptor Ionotropic Kainate 1, Human |
701 |
0.86 |
Binding ≤ 1μM
|
|
GRIK2_RAT |
P42260
|
Glutamate Receptor Ionotropic Kainate 2, Rat |
120 |
0.97 |
Binding ≤ 1μM
|
|
GRIK3_HUMAN |
Q13003
|
Glutamate Receptor Ionotropic Kainate 3, Human |
789 |
0.85 |
Binding ≤ 1μM
|
|
GRIK3_RAT |
P42264
|
Glutamate Receptor Ionotropic Kainate 3, Rat |
120 |
0.97 |
Binding ≤ 1μM
|
|
GRIK4_RAT |
Q01812
|
Glutamate Receptor Ionotropic Kainate 4, Rat |
120 |
0.97 |
Binding ≤ 1μM
|
|
GRIK5_RAT |
Q63273
|
Glutamate Receptor Ionotropic Kainate 5, Rat |
120 |
0.97 |
Binding ≤ 1μM
|
|
GRIK5_HUMAN |
Q16478
|
Glutamate Receptor Ionotropic Kainate 5, Human |
750 |
0.86 |
Binding ≤ 1μM
|
|
GRIA1_MOUSE |
P23818
|
Glutamate Receptor Ionotropic, AMPA 1, Mouse |
150 |
0.96 |
Binding ≤ 1μM
|
|
GRIA2_RAT |
P19491
|
Glutamate Receptor Ionotropic, AMPA 2, Rat |
150 |
0.96 |
Binding ≤ 1μM
|
|
GRIA2_HUMAN |
P42262
|
Glutamate Receptor Ionotropic, AMPA 2, Human |
940 |
0.84 |
Binding ≤ 1μM
|
|
GRIA3_RAT |
P19492
|
Glutamate Receptor Ionotropic, AMPA 3, Rat |
150 |
0.96 |
Binding ≤ 1μM
|
|
GRIA4_RAT |
P19493
|
Glutamate Receptor Ionotropic, AMPA 4, Rat |
150 |
0.96 |
Binding ≤ 1μM
|
|
GRIA4_HUMAN |
P48058
|
Glutamate Receptor Ionotropic, AMPA 4, Human |
868 |
0.85 |
Binding ≤ 1μM
|
|
NMDE1_RAT |
Q00959
|
Glutamate [NMDA] Receptor Subunit Epsilon 1, Rat |
200 |
0.94 |
Binding ≤ 1μM
|
|
NMDE2_RAT |
Q00960
|
Glutamate [NMDA] Receptor Subunit Epsilon 2, Rat |
200 |
0.94 |
Binding ≤ 1μM
|
|
NMDE3_RAT |
Q00961
|
Glutamate [NMDA] Receptor Subunit Epsilon 3, Rat |
200 |
0.94 |
Binding ≤ 1μM
|
|
NMDE4_RAT |
Q62645
|
Glutamate [NMDA] Receptor Subunit Epsilon 4, Rat |
200 |
0.94 |
Binding ≤ 1μM
|
|
GRM1_HUMAN |
Q13255
|
Metabotropic Glutamate Receptor 1, Human |
340 |
0.91 |
Binding ≤ 1μM
|
|
GRM2_HUMAN |
Q14416
|
Metabotropic Glutamate Receptor 2, Human |
290 |
0.92 |
Binding ≤ 1μM
|
|
Z104302 |
Z104302
|
Glutamate NMDA Receptor |
70 |
1.00 |
Binding ≤ 10μM
|
|
MURI_HELPY |
P56068
|
Glutamate Racemase, Helpy |
5800 |
0.73 |
Binding ≤ 10μM
|
|
GRIK1_RAT |
P22756
|
Glutamate Receptor Ionotropic Kainate 1, Rat |
120 |
0.97 |
Binding ≤ 10μM
|
|
GRIK1_HUMAN |
P39086
|
Glutamate Receptor Ionotropic Kainate 1, Human |
701 |
0.86 |
Binding ≤ 10μM
|
|
GRIK2_RAT |
P42260
|
Glutamate Receptor Ionotropic Kainate 2, Rat |
120 |
0.97 |
Binding ≤ 10μM
|
|
GRIK2_HUMAN |
Q13002
|
Glutamate Receptor Ionotropic Kainate 2, Human |
1106 |
0.83 |
Binding ≤ 10μM
|
|
GRIK3_RAT |
P42264
|
Glutamate Receptor Ionotropic Kainate 3, Rat |
120 |
0.97 |
Binding ≤ 10μM
|
|
GRIK3_HUMAN |
Q13003
|
Glutamate Receptor Ionotropic Kainate 3, Human |
789 |
0.85 |
Binding ≤ 10μM
|
|
GRIK4_RAT |
Q01812
|
Glutamate Receptor Ionotropic Kainate 4, Rat |
120 |
0.97 |
Binding ≤ 10μM
|
|
GRIK5_RAT |
Q63273
|
Glutamate Receptor Ionotropic Kainate 5, Rat |
120 |
0.97 |
Binding ≤ 10μM
|
|
GRIK5_HUMAN |
Q16478
|
Glutamate Receptor Ionotropic Kainate 5, Human |
750 |
0.86 |
Binding ≤ 10μM
|
|
GRIA1_HUMAN |
P42261
|
Glutamate Receptor Ionotropic, AMPA 1, Human |
1362 |
0.82 |
Binding ≤ 10μM
|
|
GRIA1_MOUSE |
P23818
|
Glutamate Receptor Ionotropic, AMPA 1, Mouse |
150 |
0.96 |
Binding ≤ 10μM
|
|
GRIA2_HUMAN |
P42262
|
Glutamate Receptor Ionotropic, AMPA 2, Human |
940 |
0.84 |
Binding ≤ 10μM
|
|
GRIA2_RAT |
P19491
|
Glutamate Receptor Ionotropic, AMPA 2, Rat |
150 |
0.96 |
Binding ≤ 10μM
|
|
GRIA3_RAT |
P19492
|
Glutamate Receptor Ionotropic, AMPA 3, Rat |
150 |
0.96 |
Binding ≤ 10μM
|
|
GRIA4_RAT |
P19493
|
Glutamate Receptor Ionotropic, AMPA 4, Rat |
150 |
0.96 |
Binding ≤ 10μM
|
|
GRIA4_HUMAN |
P48058
|
Glutamate Receptor Ionotropic, AMPA 4, Human |
868 |
0.85 |
Binding ≤ 10μM
|
|
NMDE1_RAT |
Q00959
|
Glutamate [NMDA] Receptor Subunit Epsilon 1, Rat |
200 |
0.94 |
Binding ≤ 10μM
|
|
NMDE2_RAT |
Q00960
|
Glutamate [NMDA] Receptor Subunit Epsilon 2, Rat |
200 |
0.94 |
Binding ≤ 10μM
|
|
NMDE3_RAT |
Q00961
|
Glutamate [NMDA] Receptor Subunit Epsilon 3, Rat |
200 |
0.94 |
Binding ≤ 10μM
|
|
NMDE4_RAT |
Q62645
|
Glutamate [NMDA] Receptor Subunit Epsilon 4, Rat |
200 |
0.94 |
Binding ≤ 10μM
|
|
GRM1_HUMAN |
Q13255
|
Metabotropic Glutamate Receptor 1, Human |
340 |
0.91 |
Binding ≤ 10μM
|
|
GRM1_RAT |
P23385
|
Metabotropic Glutamate Receptor 1, Rat |
10000 |
0.70 |
Binding ≤ 10μM
|
|
GRM2_HUMAN |
Q14416
|
Metabotropic Glutamate Receptor 2, Human |
1200 |
0.83 |
Binding ≤ 10μM
|
|
GRM2_RAT |
P31421
|
Metabotropic Glutamate Receptor 2, Rat |
4000 |
0.76 |
Binding ≤ 10μM
|
|
GRM3_HUMAN |
Q14832
|
Metabotropic Glutamate Receptor 3, Human |
1900 |
0.80 |
Binding ≤ 10μM
|
|
GRM3_RAT |
P31422
|
Metabotropic Glutamate Receptor 3, Rat |
9000 |
0.71 |
Binding ≤ 10μM
|
|
GRM4_HUMAN |
Q14833
|
Metabotropic Glutamate Receptor 4, Human |
1600 |
0.81 |
Binding ≤ 10μM
|
|
GRM5_HUMAN |
P41594
|
Metabotropic Glutamate Receptor 5, Human |
3100 |
0.77 |
Binding ≤ 10μM
|
|
GRM5_RAT |
P31424
|
Metabotropic Glutamate Receptor 5, Rat |
4400 |
0.75 |
Binding ≤ 10μM
|
|
GRM6_HUMAN |
O15303
|
Metabotropic Glutamate Receptor 6, Human |
4900 |
0.74 |
Binding ≤ 10μM
|
|
GRIA1_RAT |
P19490
|
Glutamate Receptor Ionotropic, AMPA 1, Rat |
214 |
0.93 |
Functional ≤ 10μM
|
|
GRIA2_RAT |
P19491
|
Glutamate Receptor Ionotropic, AMPA 2, Rat |
214 |
0.93 |
Functional ≤ 10μM
|
|
GRIA3_RAT |
P19492
|
Glutamate Receptor Ionotropic, AMPA 3, Rat |
214 |
0.93 |
Functional ≤ 10μM
|
|
GRIA4_RAT |
P19493
|
Glutamate Receptor Ionotropic, AMPA 4, Rat |
214 |
0.93 |
Functional ≤ 10μM
|
|
GRM1_RAT |
P23385
|
Metabotropic Glutamate Receptor 1, Rat |
3000 |
0.77 |
Functional ≤ 10μM
|
|
GRM1_HUMAN |
Q13255
|
Metabotropic Glutamate Receptor 1, Human |
1050 |
0.84 |
Functional ≤ 10μM
|
|
GRM8_RAT |
P70579
|
Metabotropic Glutamate Receptor 8, Rat |
8000 |
0.71 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
2500 |
0.78 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.25 |
1.72 |
-74 |
3 |
5 |
-1 |
108 |
146.122 |
4 |
↓
|
|
|
|
Analogs
-
2169443
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.23 |
-1.9 |
-75.72 |
3 |
5 |
-1 |
107 |
164.112 |
4 |
↓
|
|
|
|
|
|
|
Analogs
-
2169443
-

-
2169444
-
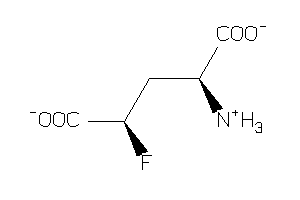
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.23 |
-1.92 |
-75.68 |
3 |
5 |
-1 |
107 |
164.112 |
4 |
↓
|
|
|
|
Analogs
-
2169445
-
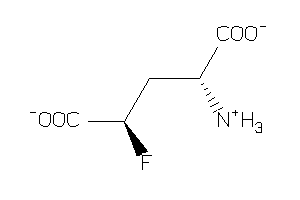
-
2169443
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.23 |
-1.89 |
-70.21 |
3 |
5 |
-1 |
107 |
164.112 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.94 |
2.83 |
-36.57 |
4 |
2 |
1 |
43 |
129.227 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.94 |
2.37 |
-0.92 |
3 |
2 |
0 |
38 |
128.219 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.06 |
2.52 |
-36.78 |
4 |
2 |
1 |
43 |
129.227 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.06 |
2.03 |
-2.43 |
3 |
2 |
0 |
38 |
128.219 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.42 |
0.53 |
-46.91 |
3 |
2 |
1 |
41 |
129.208 |
2 |
↓
|
|
|
|
|
|
|
Analogs
-
4899796
-
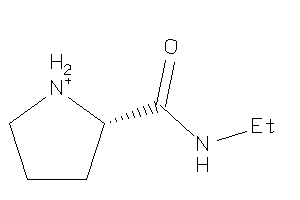
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.41 |
-0.09 |
-38.75 |
3 |
3 |
1 |
46 |
129.183 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
-0.41 |
-1.38 |
-10.92 |
2 |
3 |
0 |
41 |
128.175 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.75 |
-0.9 |
-48.42 |
4 |
3 |
1 |
60 |
129.183 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.75 |
-0.9 |
-48.41 |
4 |
3 |
1 |
60 |
129.183 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.22 |
5.65 |
-45.29 |
0 |
2 |
-1 |
40 |
163.624 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
2.22 |
4.53 |
-7.56 |
1 |
2 |
0 |
37 |
164.632 |
6 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.57 |
2.02 |
-42.07 |
4 |
2 |
1 |
40 |
165.26 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.38 |
6.28 |
-38.88 |
1 |
2 |
1 |
17 |
165.26 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.38 |
6.56 |
-81.8 |
2 |
2 |
2 |
19 |
166.268 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.02 |
3.71 |
-44.4 |
3 |
2 |
1 |
31 |
165.26 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.09 |
3.63 |
-42 |
3 |
2 |
1 |
31 |
165.26 |
2 |
↓
|
|
|
|
Analogs
-
22198608
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.88 |
3.74 |
-44.45 |
3 |
2 |
1 |
41 |
165.26 |
4 |
↓
|
|
Lo
Low (pH 4.5-6)
|
0.88 |
4.18 |
-93.33 |
4 |
2 |
2 |
42 |
166.268 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.09 |
3.64 |
-42.16 |
3 |
2 |
1 |
31 |
165.26 |
2 |
↓
|
|
|
|
Analogs
-
32220755
-
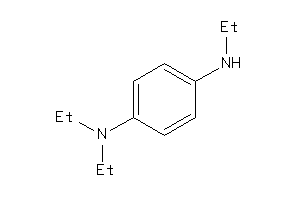
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.87 |
4.75 |
-15.85 |
3 |
2 |
0 |
30 |
165.26 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.87 |
4.67 |
-3 |
2 |
2 |
0 |
29 |
164.252 |
3 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
44651726
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.28 |
4.47 |
-117.81 |
4 |
2 |
2 |
33 |
166.268 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.28 |
3.05 |
-39.07 |
3 |
2 |
1 |
29 |
165.26 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.06 |
1.7 |
-37.3 |
5 |
2 |
1 |
54 |
165.26 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.06 |
1.21 |
-1.73 |
4 |
2 |
0 |
52 |
164.252 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.06 |
2 |
-124.85 |
6 |
2 |
2 |
55 |
166.268 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.06 |
1.88 |
-39.8 |
5 |
2 |
1 |
54 |
165.26 |
3 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.06 |
1.52 |
-2.02 |
4 |
2 |
0 |
52 |
164.252 |
3 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.06 |
2.07 |
-125.33 |
6 |
2 |
2 |
55 |
166.268 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.06 |
1.78 |
-101.13 |
6 |
2 |
2 |
55 |
166.268 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.71 |
-1.08 |
-48.9 |
0 |
2 |
-1 |
40 |
163.243 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.53 |
1.96 |
-52.95 |
3 |
2 |
1 |
41 |
165.241 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
1.53 |
1.56 |
-6.38 |
2 |
2 |
0 |
39 |
164.233 |
1 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.37 |
4.72 |
-46.59 |
1 |
3 |
1 |
32 |
165.216 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.73 |
2.62 |
-31.54 |
4 |
3 |
1 |
61 |
165.216 |
3 |
↓
|
|