|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
7.65 |
12.42 |
-6.87 |
1 |
3 |
0 |
47 |
328.537 |
19 |
↓
|
|
|
|
Analogs
-
1561822
-
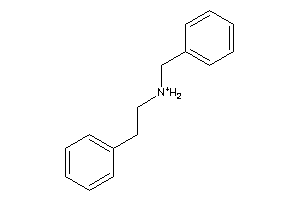
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.30 |
7.68 |
-2.95 |
1 |
1 |
0 |
12 |
211.308 |
5 |
↓
|
|
|
|
Analogs
-
585979
-
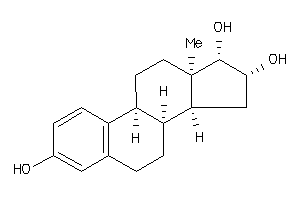
-
1736208
-
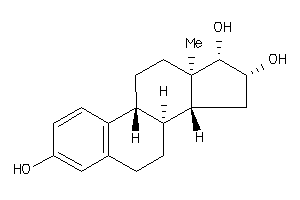
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.51 |
2.81 |
-7.02 |
3 |
3 |
0 |
61 |
288.387 |
0 |
↓
|
|
|
|
Analogs
-
6920409
-
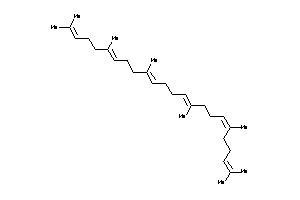
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
9.62 |
21.59 |
-0.82 |
0 |
0 |
0 |
0 |
410.73 |
15 |
↓
|
|
|
|
Analogs
-
6920409
-

Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
9.62 |
21.67 |
-1.04 |
0 |
0 |
0 |
0 |
410.73 |
15 |
↓
|
|
|
|
|
|
|
Analogs
-
6920409
-

-
1531533
-
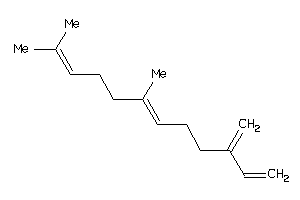
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
9.62 |
21.69 |
-1.09 |
0 |
0 |
0 |
0 |
410.73 |
15 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.74 |
2.66 |
-19.03 |
2 |
6 |
0 |
88 |
297.294 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.86 |
9.46 |
-14.45 |
0 |
3 |
0 |
28 |
278.335 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ACES-10-E |
Acetylcholinesterase (cluster #10 Of 12), Eukaryotic |
Eukaryotes |
5 |
1.66 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.39 |
5.16 |
-1.88 |
0 |
0 |
0 |
0 |
92.141 |
0 |
↓
|
|
|
|
Analogs
-
3947513
-
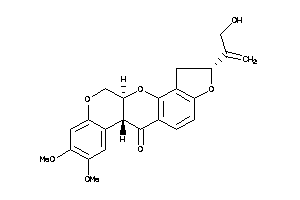
-
3947514
-
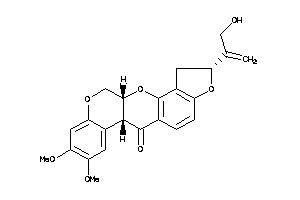
-
3947515
-
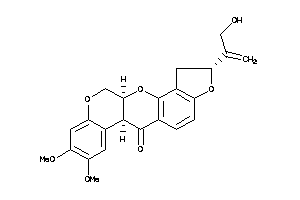
-
3947516
-
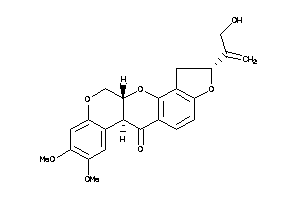
-
3978464
-
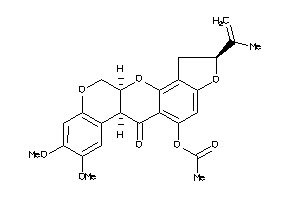
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
DUOX2-2-E |
Thyroid Oxidase 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5 |
0.40 |
Binding ≤ 10μM
|
|
NDUA4-2-E |
NADH-ubiquinone Oxidoreductase MLRQ Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
29 |
0.36 |
Binding ≤ 10μM
|
|
NDUA6-2-E |
NADH-ubiquinone Oxidoreductase B14 Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
29 |
0.36 |
Binding ≤ 10μM
|
|
NDUA8-2-E |
NADH-ubiquinone Oxidoreductase 19 KDa Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
29 |
0.36 |
Binding ≤ 10μM
|
|
NDUAA-2-E |
NADH-ubiquinone Oxidoreductase 42 KDa Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
29 |
0.36 |
Binding ≤ 10μM
|
|
NDUAB-2-E |
NADH-ubiquinone Oxidoreductase Subunit B14.7 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
29 |
0.36 |
Binding ≤ 10μM
|
|
NDUB5-2-E |
NADH-ubiquinone Oxidoreductase SGDH Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
29 |
0.36 |
Binding ≤ 10μM
|
|
NDUB6-2-E |
NADH-ubiquinone Oxidoreductase B17 Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
29 |
0.36 |
Binding ≤ 10μM
|
|
NDUB7-2-E |
NADH-ubiquinone Oxidoreductase B18 Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
29 |
0.36 |
Binding ≤ 10μM
|
|
NDUS2-2-E |
NADH-ubiquinone Oxidoreductase 49 KDa Subunit (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
29 |
0.36 |
Binding ≤ 10μM
|
|
NDUS8-2-E |
NADH-ubiquinone Oxidoreductase 23 KDa Subunit, (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
29 |
0.36 |
Binding ≤ 10μM
|
|
NU1M-1-E |
NADH-ubiquinone Oxidoreductase Chain 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.42 |
Binding ≤ 10μM |
|
NU2M-1-E |
NADH-ubiquinone Oxidoreductase Chain 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.42 |
Binding ≤ 10μM |
|
NU3M-1-E |
NADH-ubiquinone Oxidoreductase Chain 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.42 |
Binding ≤ 10μM |
|
NU4M-1-E |
NADH-ubiquinone Oxidoreductase Chain 4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.41 |
Binding ≤ 10μM |
|
NU5M-1-E |
NADH-ubiquinone Oxidoreductase Chain 5 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.42 |
Binding ≤ 10μM |
|
NU6M-1-E |
NADH-ubiquinone Oxidoreductase Chain 6 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
2 |
0.42 |
Binding ≤ 10μM |
|
Z101775-1-O |
Drosophila (cluster #1 Of 1), Other |
Other |
10000 |
0.24 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
10000 |
0.24 |
Functional ≤ 10μM
|
|
Z50531-3-O |
Caenorhabditis Elegans (cluster #3 Of 3), Other |
Other |
2000 |
0.28 |
Functional ≤ 10μM
|
|
Z50531-1-O |
Caenorhabditis Elegans (cluster #1 Of 1), Other |
Other |
2000 |
0.28 |
ADME/T ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NDUA8_HUMAN |
P51970
|
NADH-ubiquinone Oxidoreductase 19 KDa Subunit, Human |
28.8 |
0.36 |
Binding ≤ 1μM
|
|
NDUS8_HUMAN |
O00217
|
NADH-ubiquinone Oxidoreductase 23 KDa Subunit,, Human |
28.8 |
0.36 |
Binding ≤ 1μM
|
|
NDUAA_HUMAN |
O95299
|
NADH-ubiquinone Oxidoreductase 42 KDa Subunit, Human |
28.8 |
0.36 |
Binding ≤ 1μM
|
|
NDUS2_HUMAN |
O75306
|
NADH-ubiquinone Oxidoreductase 49 KDa Subunit, Human |
28.8 |
0.36 |
Binding ≤ 1μM
|
|
NDUA6_HUMAN |
P56556
|
NADH-ubiquinone Oxidoreductase B14 Subunit, Human |
28.8 |
0.36 |
Binding ≤ 1μM
|
|
NDUB6_HUMAN |
O95139
|
NADH-ubiquinone Oxidoreductase B17 Subunit, Human |
28.8 |
0.36 |
Binding ≤ 1μM
|
|
NDUB7_HUMAN |
P17568
|
NADH-ubiquinone Oxidoreductase B18 Subunit, Human |
28.8 |
0.36 |
Binding ≤ 1μM
|
|
NU1M_HUMAN |
P03886
|
NADH-ubiquinone Oxidoreductase Chain 1, Human |
1.99526231 |
0.42 |
Binding ≤ 1μM
|
|
NU2M_HUMAN |
P03891
|
NADH-ubiquinone Oxidoreductase Chain 2, Human |
1.99526231 |
0.42 |
Binding ≤ 1μM
|
|
NU3M_HUMAN |
P03897
|
NADH-ubiquinone Oxidoreductase Chain 3, Human |
1.99526231 |
0.42 |
Binding ≤ 1μM
|
|
NU4M_HUMAN |
P03905
|
NADH-ubiquinone Oxidoreductase Chain 4, Human |
1.99526231 |
0.42 |
Binding ≤ 1μM
|
|
NU5M_HUMAN |
P03915
|
NADH-ubiquinone Oxidoreductase Chain 5, Human |
1.99526231 |
0.42 |
Binding ≤ 1μM
|
|
NU6M_HUMAN |
P03923
|
NADH-ubiquinone Oxidoreductase Chain 6, Human |
1.99526231 |
0.42 |
Binding ≤ 1μM
|
|
NDUA4_HUMAN |
O00483
|
NADH-ubiquinone Oxidoreductase MLRQ Subunit, Human |
28.8 |
0.36 |
Binding ≤ 1μM
|
|
NDUB5_HUMAN |
O43674
|
NADH-ubiquinone Oxidoreductase SGDH Subunit, Human |
28.8 |
0.36 |
Binding ≤ 1μM
|
|
NDUAB_HUMAN |
Q86Y39
|
NADH-ubiquinone Oxidoreductase Subunit B14.7, Human |
28.8 |
0.36 |
Binding ≤ 1μM
|
|
DUOX2_HUMAN |
Q9NRD8
|
Thyroid Oxidase 2, Human |
5.1 |
0.40 |
Binding ≤ 1μM
|
|
NDUA8_HUMAN |
P51970
|
NADH-ubiquinone Oxidoreductase 19 KDa Subunit, Human |
28.8 |
0.36 |
Binding ≤ 10μM
|
|
NDUS8_HUMAN |
O00217
|
NADH-ubiquinone Oxidoreductase 23 KDa Subunit,, Human |
28.8 |
0.36 |
Binding ≤ 10μM
|
|
NDUAA_HUMAN |
O95299
|
NADH-ubiquinone Oxidoreductase 42 KDa Subunit, Human |
28.8 |
0.36 |
Binding ≤ 10μM
|
|
NDUS2_HUMAN |
O75306
|
NADH-ubiquinone Oxidoreductase 49 KDa Subunit, Human |
28.8 |
0.36 |
Binding ≤ 10μM
|
|
NDUA6_HUMAN |
P56556
|
NADH-ubiquinone Oxidoreductase B14 Subunit, Human |
28.8 |
0.36 |
Binding ≤ 10μM
|
|
NDUB6_HUMAN |
O95139
|
NADH-ubiquinone Oxidoreductase B17 Subunit, Human |
28.8 |
0.36 |
Binding ≤ 10μM
|
|
NDUB7_HUMAN |
P17568
|
NADH-ubiquinone Oxidoreductase B18 Subunit, Human |
28.8 |
0.36 |
Binding ≤ 10μM
|
|
NU1M_HUMAN |
P03886
|
NADH-ubiquinone Oxidoreductase Chain 1, Human |
1.99526231 |
0.42 |
Binding ≤ 10μM
|
|
NU2M_HUMAN |
P03891
|
NADH-ubiquinone Oxidoreductase Chain 2, Human |
1.99526231 |
0.42 |
Binding ≤ 10μM
|
|
NU3M_HUMAN |
P03897
|
NADH-ubiquinone Oxidoreductase Chain 3, Human |
1.99526231 |
0.42 |
Binding ≤ 10μM
|
|
NU4M_HUMAN |
P03905
|
NADH-ubiquinone Oxidoreductase Chain 4, Human |
1.99526231 |
0.42 |
Binding ≤ 10μM
|
|
NU5M_HUMAN |
P03915
|
NADH-ubiquinone Oxidoreductase Chain 5, Human |
1.99526231 |
0.42 |
Binding ≤ 10μM
|
|
NU6M_HUMAN |
P03923
|
NADH-ubiquinone Oxidoreductase Chain 6, Human |
1.99526231 |
0.42 |
Binding ≤ 10μM
|
|
NDUA4_HUMAN |
O00483
|
NADH-ubiquinone Oxidoreductase MLRQ Subunit, Human |
28.8 |
0.36 |
Binding ≤ 10μM
|
|
NDUB5_HUMAN |
O43674
|
NADH-ubiquinone Oxidoreductase SGDH Subunit, Human |
28.8 |
0.36 |
Binding ≤ 10μM
|
|
NDUAB_HUMAN |
Q86Y39
|
NADH-ubiquinone Oxidoreductase Subunit B14.7, Human |
28.8 |
0.36 |
Binding ≤ 10μM
|
|
DUOX2_HUMAN |
Q9NRD8
|
Thyroid Oxidase 2, Human |
5.1 |
0.40 |
Binding ≤ 10μM
|
|
Z50531 |
Z50531
|
Caenorhabditis Elegans |
2000 |
0.28 |
Functional ≤ 10μM
|
|
Z101775 |
Z101775
|
Drosophila |
10000 |
0.24 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
10000 |
0.24 |
Functional ≤ 10μM
|
|
Z50531 |
Z50531
|
Caenorhabditis Elegans |
2000 |
0.28 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.59 |
8.74 |
-11.43 |
0 |
6 |
0 |
63 |
394.423 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.21 |
1.34 |
-4.5 |
0 |
1 |
0 |
12 |
181.238 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.09 |
9.89 |
-20.95 |
0 |
5 |
0 |
58 |
317.328 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.77 |
5.04 |
-13.67 |
1 |
5 |
0 |
65 |
237.255 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.11 |
1.31 |
-2.44 |
0 |
1 |
0 |
9 |
164.966 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
P2RX2-1-E |
P2X Purinoceptor 2 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.35 |
Functional ≤ 10μM
|
|
P2RX3-1-E |
P2X Purinoceptor 3 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
10000 |
0.35 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.39 |
4.67 |
-10.73 |
4 |
6 |
0 |
96 |
274.324 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.39 |
5.17 |
-38.05 |
5 |
6 |
1 |
98 |
275.332 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PGH2-4-E |
Cyclooxygenase-2 (cluster #4 Of 8), Eukaryotic |
Eukaryotes |
50 |
0.38 |
Binding ≤ 10μM
|
|
Q8SPQ9-2-E |
Cyclooxygenase-2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
630 |
0.32 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.96 |
3.89 |
-17.42 |
2 |
6 |
0 |
87 |
397.378 |
5 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.46 |
10.67 |
-35.13 |
0 |
2 |
1 |
9 |
262.398 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.94 |
9.13 |
-10.69 |
0 |
3 |
0 |
28 |
321.549 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.42 |
15.64 |
-27.12 |
0 |
1 |
1 |
4 |
290.515 |
13 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.11 |
4.9 |
-10.62 |
1 |
5 |
0 |
60 |
266.322 |
5 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.74 |
2.91 |
-45.31 |
0 |
5 |
-1 |
67 |
265.314 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.51 |
3.77 |
-23.86 |
0 |
6 |
0 |
74 |
415.593 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.84 |
12.26 |
-23.24 |
0 |
5 |
0 |
58 |
362.771 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.41 |
5.81 |
-11.67 |
1 |
9 |
0 |
120 |
457.386 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.86 |
3.21 |
-42.47 |
0 |
9 |
-1 |
123 |
456.378 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ESR2-1-E |
Estrogen Receptor Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
24 |
0.46 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.74 |
3.31 |
-7.52 |
3 |
5 |
0 |
87 |
322.401 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.74 |
4.46 |
-41.9 |
2 |
5 |
-1 |
90 |
321.393 |
0 |
↓
|
|
|
|
|
|
|
Analogs
-
17653974
-
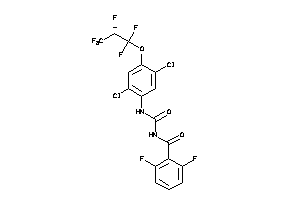
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.82 |
3.99 |
-17.88 |
2 |
5 |
0 |
67 |
511.152 |
6 |
↓
|
|
|
|
Analogs
-
8214421
-
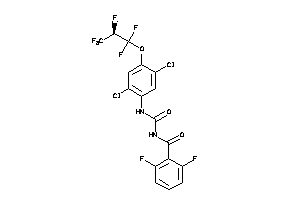
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.82 |
9.36 |
-17.54 |
2 |
5 |
0 |
67 |
511.152 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.42 |
3.18 |
-10.43 |
0 |
5 |
0 |
61 |
327.528 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.42 |
6.96 |
-10.53 |
0 |
5 |
0 |
62 |
327.528 |
8 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.67 |
3.95 |
-8.09 |
0 |
4 |
0 |
45 |
220.976 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.71 |
10.78 |
-17.36 |
1 |
6 |
0 |
78 |
361.438 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.01 |
7.11 |
-6.01 |
1 |
1 |
0 |
16 |
199.278 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.42 |
-4.51 |
-15.63 |
6 |
7 |
0 |
146 |
380.662 |
3 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.31 |
14.08 |
-17.65 |
1 |
6 |
0 |
78 |
417.546 |
15 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-12-O |
Plasmodium Falciparum (cluster #12 Of 22), Other |
Other |
9730 |
0.64 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
3370 |
0.70 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.06 |
4.17 |
-16.61 |
1 |
2 |
0 |
33 |
192.045 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.52 |
2.83 |
-36.68 |
0 |
2 |
-1 |
36 |
191.037 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.59 |
4.04 |
-5.16 |
0 |
2 |
0 |
29 |
192.045 |
0 |
↓
|
|
|
|
Analogs
-
1537480
-
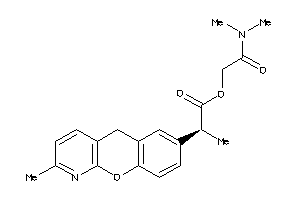
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.03 |
8.51 |
-18.41 |
0 |
6 |
0 |
69 |
354.406 |
5 |
↓
|
|
|
|
Analogs
-
5161047
-
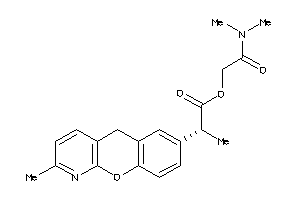
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.03 |
8.51 |
-18.41 |
0 |
6 |
0 |
69 |
354.406 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.16 |
3.96 |
-3.26 |
1 |
1 |
0 |
20 |
150.221 |
1 |
↓
|
|
|
|
Analogs
-
36175852
-
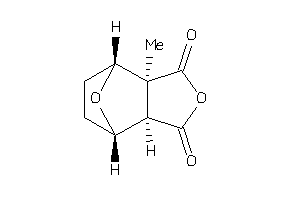
-
8674160
-
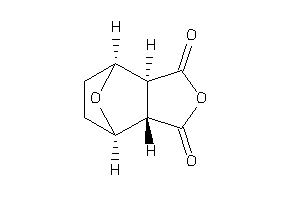
-
8143882
-
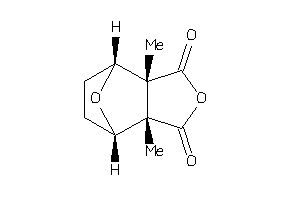
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
2A5A-1-E |
Serine/threonine Protein Phosphatase 2A, 56 KDa Regulatory Subunit, Alpha Isoform (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
600 |
0.62 |
Binding ≤ 10μM
|
|
2AAA-1-E |
Serine/threonine Protein Phosphatase 2A, 65 KDa Regulatory Subunit A, Alpha Isoform (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8000 |
0.51 |
Binding ≤ 10μM |
|
2ABG-1-E |
Serine/threonine Protein Phosphatase 2A, 55 KDa Regulatory Subunit B, Gamma Isoform (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
600 |
0.62 |
Binding ≤ 10μM
|
|
PP1A-1-E |
Serine/threonine Protein Phosphatase PP1-alpha Catalytic Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.63 |
Binding ≤ 10μM
|
|
PP1G-1-E |
Serine/threonine Protein Phosphatase PP1-gamma Catalytic Subunit (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
500 |
0.63 |
Binding ≤ 10μM
|
|
PP2AA-1-E |
Serine/threonine Protein Phosphatase 2A, Catalytic Subunit, Alpha Isoform (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
600 |
0.62 |
Binding ≤ 10μM
|
|
PP2AB-1-E |
Serine/threonine Protein Phosphatase 2A, Catalytic Subunit, Beta Isoform (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
65 |
0.72 |
Binding ≤ 10μM |
|
PPM1B-1-E |
Protein Phosphatase 2C Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
430 |
0.64 |
Binding ≤ 10μM
|
|
PPM1J-1-E |
Protein Phosphatase 2A Catalytic Subunit Zeta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
65 |
0.72 |
Binding ≤ 10μM |
|
PTN1-3-E |
Protein-tyrosine Phosphatase 1B (cluster #3 Of 4), Eukaryotic |
Eukaryotes |
3600 |
0.54 |
Binding ≤ 10μM
|
|
PTPA-1-E |
Protein Phosphatase 2A Regulatory Subunit B' (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
600 |
0.62 |
Binding ≤ 10μM
|
|
Q8AWE3-1-E |
Serine-threonine Protein Phosphatase 2A Regulatory Subunit (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
360 |
0.64 |
Binding ≤ 10μM
|
|
Z50425-1-O |
Plasmodium Falciparum (cluster #1 Of 22), Other |
Other |
9000 |
0.50 |
Functional ≤ 10μM
|
|
Z80001-2-O |
143B (cluster #2 Of 2), Other |
Other |
10000 |
0.50 |
Functional ≤ 10μM
|
|
Z80166-2-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #2 Of 12), Other |
Other |
6400 |
0.52 |
Functional ≤ 10μM
|
|
Z80928-4-O |
HCT-116 (Colon Carcinoma Cells) (cluster #4 Of 9), Other |
Other |
9000 |
0.50 |
Functional ≤ 10μM
|
|
Z81034-2-O |
A2780 (Ovarian Carcinoma Cells) (cluster #2 Of 10), Other |
Other |
10000 |
0.50 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PPM1J_MOUSE |
Q149T7
|
Protein Phosphatase 2A Catalytic Subunit Zeta, Mouse |
65 |
0.72 |
Binding ≤ 1μM
|
|
PTPA_HUMAN |
Q15257
|
Protein Phosphatase 2A Regulatory Subunit B', Human |
600 |
0.62 |
Binding ≤ 1μM
|
|
PPM1B_HUMAN |
O75688
|
Protein Phosphatase 2C Beta, Human |
430 |
0.64 |
Binding ≤ 1μM
|
|
Q8AWE3_CHICK |
Q8AWE3
|
Serine-threonine Protein Phosphatase 2A Regulatory Subunit, Chick |
200 |
0.67 |
Binding ≤ 1μM
|
|
2ABG_HUMAN |
Q9Y2T4
|
Serine/threonine Protein Phosphatase 2A, 55 KDa Regulatory Subunit B, Gamma Isoform, Human |
600 |
0.62 |
Binding ≤ 1μM
|
|
2A5A_HUMAN |
Q15172
|
Serine/threonine Protein Phosphatase 2A, 56 KDa Regulatory Subunit, Alpha Isoform, Human |
600 |
0.62 |
Binding ≤ 1μM
|
|
PP2AA_HUMAN |
P67775
|
Serine/threonine Protein Phosphatase 2A, Catalytic Subunit, Alpha Isoform, Human |
600 |
0.62 |
Binding ≤ 1μM
|
|
PP2AA_MOUSE |
P63330
|
Serine/threonine Protein Phosphatase 2A, Catalytic Subunit, Alpha Isoform, Mouse |
65 |
0.72 |
Binding ≤ 1μM
|
|
PP2AB_HUMAN |
P62714
|
Serine/threonine Protein Phosphatase 2A, Catalytic Subunit, Beta Isoform, Human |
600 |
0.62 |
Binding ≤ 1μM
|
|
PP1A_HUMAN |
P62136
|
Serine/threonine Protein Phosphatase PP1-alpha Catalytic Subunit, Human |
160 |
0.68 |
Binding ≤ 1μM
|
|
PP1G_HUMAN |
P36873
|
Serine/threonine Protein Phosphatase PP1-gamma Catalytic Subunit, Human |
160 |
0.68 |
Binding ≤ 1μM
|
|
PPM1J_MOUSE |
Q149T7
|
Protein Phosphatase 2A Catalytic Subunit Zeta, Mouse |
65 |
0.72 |
Binding ≤ 10μM
|
|
PTPA_HUMAN |
Q15257
|
Protein Phosphatase 2A Regulatory Subunit B', Human |
600 |
0.62 |
Binding ≤ 10μM
|
|
PPM1B_HUMAN |
O75688
|
Protein Phosphatase 2C Beta, Human |
1780 |
0.57 |
Binding ≤ 10μM
|
|
PTN1_HUMAN |
P18031
|
Protein-tyrosine Phosphatase 1B, Human |
3600 |
0.54 |
Binding ≤ 10μM
|
|
Q8AWE3_CHICK |
Q8AWE3
|
Serine-threonine Protein Phosphatase 2A Regulatory Subunit, Chick |
1700 |
0.58 |
Binding ≤ 10μM
|
|
2ABG_HUMAN |
Q9Y2T4
|
Serine/threonine Protein Phosphatase 2A, 55 KDa Regulatory Subunit B, Gamma Isoform, Human |
600 |
0.62 |
Binding ≤ 10μM
|
|
2A5A_HUMAN |
Q15172
|
Serine/threonine Protein Phosphatase 2A, 56 KDa Regulatory Subunit, Alpha Isoform, Human |
600 |
0.62 |
Binding ≤ 10μM
|
|
2AAA_MOUSE |
Q76MZ3
|
Serine/threonine Protein Phosphatase 2A, 65 KDa Regulatory Subunit A, Alpha Isoform, Mouse |
8000 |
0.51 |
Binding ≤ 10μM
|
|
PP2AA_HUMAN |
P67775
|
Serine/threonine Protein Phosphatase 2A, Catalytic Subunit, Alpha Isoform, Human |
600 |
0.62 |
Binding ≤ 10μM
|
|
PP2AA_MOUSE |
P63330
|
Serine/threonine Protein Phosphatase 2A, Catalytic Subunit, Alpha Isoform, Mouse |
65 |
0.72 |
Binding ≤ 10μM
|
|
PP2AB_HUMAN |
P62714
|
Serine/threonine Protein Phosphatase 2A, Catalytic Subunit, Beta Isoform, Human |
600 |
0.62 |
Binding ≤ 10μM
|
|
PP1A_HUMAN |
P62136
|
Serine/threonine Protein Phosphatase PP1-alpha Catalytic Subunit, Human |
160 |
0.68 |
Binding ≤ 10μM
|
|
PP1G_HUMAN |
P36873
|
Serine/threonine Protein Phosphatase PP1-gamma Catalytic Subunit, Human |
160 |
0.68 |
Binding ≤ 10μM
|
|
Z80001 |
Z80001
|
143B |
10000 |
0.50 |
Functional ≤ 10μM
|
|
Z81034 |
Z81034
|
A2780 (Ovarian Carcinoma Cells) |
10000 |
0.50 |
Functional ≤ 10μM
|
|
Z80928 |
Z80928
|
HCT-116 (Colon Carcinoma Cells) |
9000 |
0.50 |
Functional ≤ 10μM
|
|
Z80166 |
Z80166
|
HT-29 (Colon Adenocarcinoma Cells) |
6400 |
0.52 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
2511.88643 |
0.56 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.37 |
4.64 |
-10.95 |
0 |
4 |
0 |
53 |
196.202 |
0 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.76 |
6.96 |
-6.82 |
1 |
2 |
0 |
25 |
264.419 |
3 |
↓
|
|
|
|
Analogs
-
1532777
-
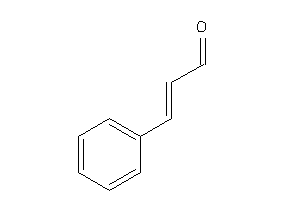
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.48 |
5.34 |
-6.27 |
0 |
1 |
0 |
17 |
132.162 |
2 |
↓
|
|