|
|
Analogs
-
2581651
-
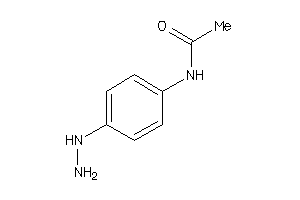
-
34396225
-
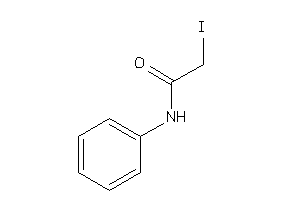
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.16 |
3.39 |
-8.79 |
1 |
2 |
0 |
29 |
135.166 |
1 |
↓
|
|
|
|
Analogs
-
4217364
-
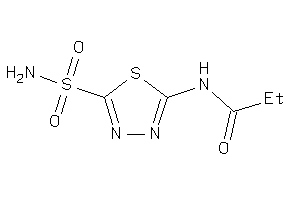
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH-1-A |
Carbonic Anhydrase (cluster #1 Of 2), Archaea |
Archaea |
60 |
0.78 |
Binding ≤ 10μM
|
|
CYNT-2-B |
Carbonic Anhydrase (cluster #2 Of 3), Bacterial |
Bacteria |
9 |
0.87 |
Binding ≤ 10μM
|
|
P96878-1-B |
PROBABLE TRANSMEMBRANE CARBONIC ANHYDRASE (CARBONATE DEHYDRATASE) (CARBONIC DEHYDRATASE) (cluster #1 Of 2), Bacterial |
Bacteria |
12 |
0.85 |
Binding ≤ 10μM
|
|
Y1284-1-B |
Uncharacterized Protein Rv1284/MT1322 (cluster #1 Of 2), Bacterial |
Bacteria |
481 |
0.68 |
Binding ≤ 10μM
|
|
B5SU02-5-E |
Alpha Carbonic Anhydrase (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
16 |
0.84 |
Binding ≤ 10μM
|
|
C0IX24-4-E |
Carbonic Anhydrase (cluster #4 Of 5), Eukaryotic |
Eukaryotes |
74 |
0.77 |
Binding ≤ 10μM
|
|
CAH1-10-E |
Carbonic Anhydrase I (cluster #10 Of 12), Eukaryotic |
Eukaryotes |
900 |
0.65 |
Binding ≤ 10μM
|
|
CAH10-2-E |
Carbonic Anhydrase-related Protein 10 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
440 |
0.68 |
Binding ≤ 10μM
|
|
CAH11-2-E |
Carbonic Anhydrase-related Protein 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
440 |
0.68 |
Binding ≤ 10μM
|
|
CAH12-8-E |
Carbonic Anhydrase XII (cluster #8 Of 9), Eukaryotic |
Eukaryotes |
6 |
0.89 |
Binding ≤ 10μM |
|
CAH13-2-E |
Carbonic Anhydrase XIII (cluster #2 Of 7), Eukaryotic |
Eukaryotes |
490 |
0.68 |
Binding ≤ 10μM
|
|
CAH14-7-E |
Carbonic Anhydrase XIV (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
440 |
0.68 |
Binding ≤ 10μM
|
|
CAH15-6-E |
Carbonic Anhydrase 15 (cluster #6 Of 6), Eukaryotic |
Eukaryotes |
72 |
0.77 |
Binding ≤ 10μM
|
|
CAH2-11-E |
Carbonic Anhydrase II (cluster #11 Of 15), Eukaryotic |
Eukaryotes |
7 |
0.88 |
Binding ≤ 10μM
|
|
CAH3-5-E |
Carbonic Anhydrase III (cluster #5 Of 6), Eukaryotic |
Eukaryotes |
7 |
0.88 |
Binding ≤ 10μM
|
|
CAH4-12-E |
Carbonic Anhydrase IV (cluster #12 Of 16), Eukaryotic |
Eukaryotes |
72 |
0.77 |
Binding ≤ 10μM
|
|
CAH5A-2-E |
Carbonic Anhydrase VA (cluster #2 Of 10), Eukaryotic |
Eukaryotes |
7 |
0.88 |
Binding ≤ 10μM
|
|
CAH5B-2-E |
Carbonic Anhydrase VB (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
54 |
0.78 |
Binding ≤ 10μM |
|
CAH6-1-E |
Carbonic Anhydrase VI (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
7 |
0.88 |
Binding ≤ 10μM
|
|
CAH7-7-E |
Carbonic Anhydrase VII (cluster #7 Of 8), Eukaryotic |
Eukaryotes |
16 |
0.84 |
Binding ≤ 10μM
|
|
CAH8-2-E |
Carbonic Anhydrase-related Protein 8 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
440 |
0.68 |
Binding ≤ 10μM
|
|
CAH9-10-E |
Carbonic Anhydrase IX (cluster #10 Of 11), Eukaryotic |
Eukaryotes |
1000 |
0.65 |
Binding ≤ 10μM
|
|
Q2PCB5-1-E |
Carbonic Anhydrase (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
5720 |
0.56 |
Binding ≤ 10μM |
|
CAH1-3-E |
Carbonic Anhydrase I (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
250 |
0.71 |
Functional ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.92 |
Functional ≤ 10μM
|
|
CAH4-1-E |
Carbonic Anhydrase IV (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
70 |
0.77 |
Functional ≤ 10μM
|
|
CAH9-1-E |
Carbonic Anhydrase IX (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
25 |
0.82 |
Functional ≤ 10μM
|
|
CAN-3-F |
Carbonic Anhydrase (cluster #3 Of 3), Fungal |
Fungi |
83 |
0.76 |
Binding ≤ 10μM
|
|
Q3I4V7-2-F |
Carbonic Anhydrase 2 (cluster #2 Of 4), Fungal |
Fungi |
10 |
0.86 |
Binding ≤ 10μM
|
|
Q5AJ71-3-F |
Carbonic Anhydrase (cluster #3 Of 4), Fungal |
Fungi |
40 |
0.80 |
Binding ≤ 10μM
|
|
Q6FTL6-1-F |
Carbonic Anhydrase (cluster #1 Of 1), Fungal |
Fungi |
11 |
0.86 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
B5SU02_9CNID |
B5SU02
|
Alpha Carbonic Anhydrase, 9cnid |
16 |
0.84 |
Binding ≤ 1μM
|
|
CYNT_MYCTU |
O53573
|
Carbonic Anhydrase, Myctu |
9 |
0.87 |
Binding ≤ 1μM
|
|
C0IX24_9CNID |
C0IX24
|
Carbonic Anhydrase, 9cnid |
74 |
0.77 |
Binding ≤ 1μM
|
|
CAN_YEAST |
P53615
|
Carbonic Anhydrase, Yeast |
82.6 |
0.76 |
Binding ≤ 1μM
|
|
CAH_METTE |
P40881
|
Carbonic Anhydrase, Mette |
60 |
0.78 |
Binding ≤ 1μM
|
|
Q6FTL6_CANGA |
Q6FTL6
|
Carbonic Anhydrase, Canga |
11 |
0.86 |
Binding ≤ 1μM
|
|
Q5AJ71_CANAL |
Q5AJ71
|
Carbonic Anhydrase, Canal |
130 |
0.74 |
Binding ≤ 1μM
|
|
CYNT_HELPY |
O24855
|
Carbonic Anhydrase 1, Helpy |
20 |
0.83 |
Binding ≤ 1μM
|
|
CAH15_MOUSE |
Q99N23
|
Carbonic Anhydrase 15, Mouse |
72 |
0.77 |
Binding ≤ 1μM
|
|
Q3I4V7_CRYNV |
Q3I4V7
|
Carbonic Anhydrase 2, Crynv |
10 |
0.86 |
Binding ≤ 1μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH2_RAT |
P27139
|
Carbonic Anhydrase II, Rat |
12 |
0.85 |
Binding ≤ 1μM
|
|
CAH2_BOVIN |
P00921
|
Carbonic Anhydrase II, Bovin |
16 |
0.84 |
Binding ≤ 1μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH3_RAT |
P14141
|
Carbonic Anhydrase III, Rat |
12 |
0.85 |
Binding ≤ 1μM
|
|
CAH3_HUMAN |
P07451
|
Carbonic Anhydrase III, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH4_RAT |
P48284
|
Carbonic Anhydrase IV, Rat |
12 |
0.85 |
Binding ≤ 1μM
|
|
CAH4_BOVIN |
Q95323
|
Carbonic Anhydrase IV, Bovin |
120 |
0.75 |
Binding ≤ 1μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH5A_MOUSE |
P23589
|
Carbonic Anhydrase VA, Mouse |
58 |
0.78 |
Binding ≤ 1μM
|
|
CAH5A_RAT |
P43165
|
Carbonic Anhydrase VA, Rat |
12 |
0.85 |
Binding ≤ 1μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH5B_MOUSE |
Q9QZA0
|
Carbonic Anhydrase VB, Mouse |
58 |
0.78 |
Binding ≤ 1μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH6_BOVIN |
P18915
|
Carbonic Anhydrase VI, Bovin |
16 |
0.84 |
Binding ≤ 1μM
|
|
CAH7_MOUSE |
Q9ERQ8
|
Carbonic Anhydrase VII, Mouse |
16 |
0.84 |
Binding ≤ 1μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH13_MOUSE |
Q9D6N1
|
Carbonic Anhydrase XIII, Mouse |
17 |
0.84 |
Binding ≤ 1μM
|
|
CAH13_HUMAN |
Q8N1Q1
|
Carbonic Anhydrase XIII, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH10_HUMAN |
Q9NS85
|
Carbonic Anhydrase-related Protein 10, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH11_HUMAN |
O75493
|
Carbonic Anhydrase-related Protein 2, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
CAH8_HUMAN |
P35219
|
Carbonic Anhydrase-related Protein 8, Human |
0.8 |
0.98 |
Binding ≤ 1μM
|
|
P96878_MYCTU |
P96878
|
PROBABLE TRANSMEMBRANE CARBONIC ANHYDRASE (CARBONATE DEHYDRATASE) (CARBONIC DEHYDRATASE), Myctu |
100 |
0.75 |
Binding ≤ 1μM
|
|
Y1284_MYCTU |
P64797
|
Uncharacterized Protein Rv1284/MT1322, Myctu |
250 |
0.71 |
Binding ≤ 1μM
|
|
B5SU02_9CNID |
B5SU02
|
Alpha Carbonic Anhydrase, 9cnid |
16 |
0.84 |
Binding ≤ 10μM
|
|
C0IX24_9CNID |
C0IX24
|
Carbonic Anhydrase, 9cnid |
74 |
0.77 |
Binding ≤ 10μM
|
|
CAN_YEAST |
P53615
|
Carbonic Anhydrase, Yeast |
82.6 |
0.76 |
Binding ≤ 10μM
|
|
Q6FTL6_CANGA |
Q6FTL6
|
Carbonic Anhydrase, Canga |
11 |
0.86 |
Binding ≤ 10μM
|
|
CAH_METTE |
P40881
|
Carbonic Anhydrase, Mette |
1430 |
0.63 |
Binding ≤ 10μM
|
|
Q5AJ71_CANAL |
Q5AJ71
|
Carbonic Anhydrase, Canal |
130 |
0.74 |
Binding ≤ 10μM
|
|
Q2PCB5_DICLA |
Q2PCB5
|
Carbonic Anhydrase, Dicla |
5720 |
0.56 |
Binding ≤ 10μM |
|
CYNT_MYCTU |
O53573
|
Carbonic Anhydrase, Myctu |
9 |
0.87 |
Binding ≤ 10μM
|
|
CYNT_HELPY |
O24855
|
Carbonic Anhydrase 1, Helpy |
20 |
0.83 |
Binding ≤ 10μM
|
|
CAH15_MOUSE |
Q99N23
|
Carbonic Anhydrase 15, Mouse |
72 |
0.77 |
Binding ≤ 10μM
|
|
Q3I4V7_CRYNV |
Q3I4V7
|
Carbonic Anhydrase 2, Crynv |
10 |
0.86 |
Binding ≤ 10μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH2_BOVIN |
P00921
|
Carbonic Anhydrase II, Bovin |
16 |
0.84 |
Binding ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH2_RAT |
P27139
|
Carbonic Anhydrase II, Rat |
12 |
0.85 |
Binding ≤ 10μM
|
|
CAH3_RAT |
P14141
|
Carbonic Anhydrase III, Rat |
12 |
0.85 |
Binding ≤ 10μM
|
|
CAH3_HUMAN |
P07451
|
Carbonic Anhydrase III, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH4_RAT |
P48284
|
Carbonic Anhydrase IV, Rat |
12 |
0.85 |
Binding ≤ 10μM
|
|
CAH4_BOVIN |
Q95323
|
Carbonic Anhydrase IV, Bovin |
120 |
0.75 |
Binding ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH5A_RAT |
P43165
|
Carbonic Anhydrase VA, Rat |
12 |
0.85 |
Binding ≤ 10μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH5A_MOUSE |
P23589
|
Carbonic Anhydrase VA, Mouse |
58 |
0.78 |
Binding ≤ 10μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH5B_MOUSE |
Q9QZA0
|
Carbonic Anhydrase VB, Mouse |
58 |
0.78 |
Binding ≤ 10μM
|
|
CAH6_BOVIN |
P18915
|
Carbonic Anhydrase VI, Bovin |
16 |
0.84 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH7_MOUSE |
Q9ERQ8
|
Carbonic Anhydrase VII, Mouse |
16 |
0.84 |
Binding ≤ 10μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH13_MOUSE |
Q9D6N1
|
Carbonic Anhydrase XIII, Mouse |
1050 |
0.64 |
Binding ≤ 10μM
|
|
CAH13_HUMAN |
Q8N1Q1
|
Carbonic Anhydrase XIII, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH14_HUMAN |
Q9ULX7
|
Carbonic Anhydrase XIV, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH10_HUMAN |
Q9NS85
|
Carbonic Anhydrase-related Protein 10, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH11_HUMAN |
O75493
|
Carbonic Anhydrase-related Protein 2, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
CAH8_HUMAN |
P35219
|
Carbonic Anhydrase-related Protein 8, Human |
0.8 |
0.98 |
Binding ≤ 10μM
|
|
P96878_MYCTU |
P96878
|
PROBABLE TRANSMEMBRANE CARBONIC ANHYDRASE (CARBONATE DEHYDRATASE) (CARBONIC DEHYDRATASE), Myctu |
100 |
0.75 |
Binding ≤ 10μM
|
|
Y1284_MYCTU |
P64797
|
Uncharacterized Protein Rv1284/MT1322, Myctu |
250 |
0.71 |
Binding ≤ 10μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
250 |
0.71 |
Functional ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
12 |
0.85 |
Functional ≤ 10μM
|
|
CAH4_BOVIN |
Q95323
|
Carbonic Anhydrase IV, Bovin |
70 |
0.77 |
Functional ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
25 |
0.82 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.15 |
-4.36 |
-19.16 |
3 |
7 |
0 |
115 |
222.251 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
-1.15 |
-3.88 |
-51.46 |
2 |
7 |
-1 |
113 |
221.243 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.44 |
1.78 |
-47.64 |
1 |
4 |
-1 |
69 |
162.19 |
3 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.34 |
13.46 |
-81.52 |
1 |
4 |
0 |
57 |
348.446 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.34 |
11 |
-54.83 |
0 |
4 |
-1 |
56 |
347.438 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.34 |
13.76 |
-90.84 |
2 |
4 |
1 |
59 |
349.454 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z101794-1-O |
Felid Herpesvirus 1 (cluster #1 Of 1), Other |
Other |
1000 |
0.53 |
Functional ≤ 10μM |
|
Z50515-2-O |
Human Herpesvirus 2 (cluster #2 Of 2), Other |
Other |
9600 |
0.44 |
Functional ≤ 10μM
|
|
Z50517-1-O |
Human Herpesvirus 5 Strain AD169 (cluster #1 Of 2), Other |
Other |
7600 |
0.45 |
Functional ≤ 10μM
|
|
Z50518-3-O |
Human Herpesvirus 4 (cluster #3 Of 5), Other |
Other |
6900 |
0.45 |
Functional ≤ 10μM
|
|
Z50527-1-O |
Human Herpesvirus 3 (cluster #1 Of 3), Other |
Other |
900 |
0.53 |
Functional ≤ 10μM
|
|
Z50527-3-O |
Human Herpesvirus 3 (cluster #3 Of 3), Other |
Other |
4900 |
0.46 |
Functional ≤ 10μM |
|
Z50530-1-O |
Human Herpesvirus 5 (cluster #1 Of 5), Other |
Other |
7500 |
0.45 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
90 |
0.62 |
Functional ≤ 10μM
|
|
Z50600-1-O |
Vaccinia Virus (cluster #1 Of 2), Other |
Other |
384 |
0.56 |
Functional ≤ 10μM
|
|
Z50602-2-O |
Human Herpesvirus 1 (cluster #2 Of 5), Other |
Other |
990 |
0.53 |
Functional ≤ 10μM
|
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
20 |
0.67 |
Functional ≤ 10μM
|
|
Z80040-1-O |
BHK-21 (Kidney Cells) (cluster #1 Of 1), Other |
Other |
2000 |
0.50 |
Functional ≤ 10μM
|
|
Z80291-2-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
5000 |
0.46 |
Functional ≤ 10μM
|
|
Z80414-1-O |
Raji (B-lymphoblastic Cells) (cluster #1 Of 3), Other |
Other |
2900 |
0.48 |
Functional ≤ 10μM
|
|
Z80583-7-O |
Vero (Kidney Cells) (cluster #7 Of 7), Other |
Other |
7540 |
0.45 |
Functional ≤ 10μM
|
|
Z80942-1-O |
HEL (Embryonic Lung Cells) (cluster #1 Of 2), Other |
Other |
900 |
0.53 |
Functional ≤ 10μM
|
|
Z80954-1-O |
HFF (Foreskin Fibroblasts) (cluster #1 Of 4), Other |
Other |
8100 |
0.45 |
Functional ≤ 10μM
|
|
Z81247-5-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #5 Of 9), Other |
Other |
8710 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.61 |
-0.6 |
-19.69 |
4 |
8 |
0 |
119 |
225.208 |
4 |
↓
|
|
|
|
Analogs
-
2169830
-
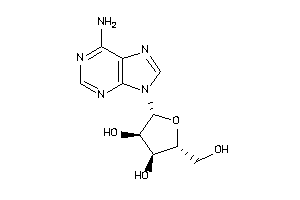
-
3201876
-
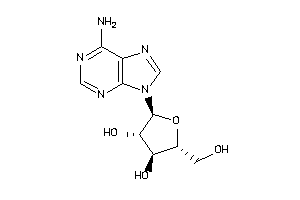
-
3201878
-
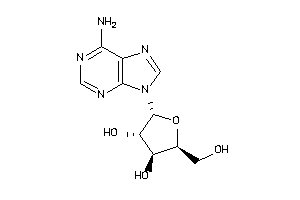
-
3978047
-
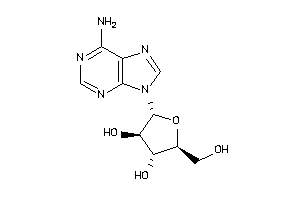
-
3978049
-
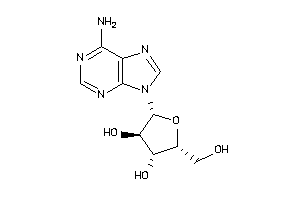
Draw
Identity
99%
90%
80%
70%
Notice: Undefined index: field_name in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 244
Notice: Undefined index: synonym in /domains/zinc12/htdocs/lib/zinc/reporter/ZincAuxiliaryInfoReports.php on line 245
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.85 |
-5.31 |
-13.62 |
5 |
9 |
0 |
140 |
267.245 |
2 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
1730540
-
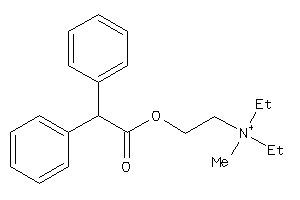
-
2003680
-
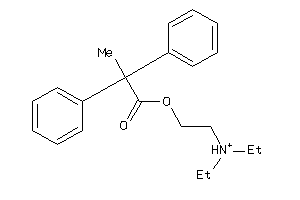
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.15 |
12.85 |
-36.39 |
1 |
3 |
1 |
31 |
312.433 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.92 |
5.45 |
-8.46 |
2 |
5 |
0 |
70 |
265.338 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
2.92 |
5.45 |
-9.77 |
2 |
5 |
0 |
70 |
265.338 |
4 |
↓
|
|
|
|
Analogs
-
44125733
-
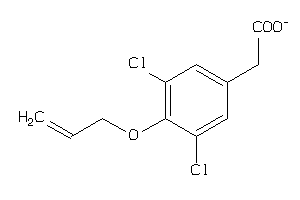
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.67 |
7.19 |
-44.05 |
0 |
3 |
-1 |
49 |
225.651 |
5 |
↓
|
|
|
|
Analogs
-
5820938
-
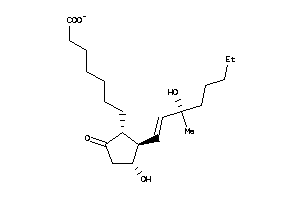
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PE2R1-2-E |
Prostanoid EP1 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
6 |
0.46 |
Binding ≤ 10μM
|
|
PE2R2-2-E |
Prostanoid EP2 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
41 |
0.41 |
Binding ≤ 10μM
|
|
PE2R3-2-E |
Prostanoid EP3 Receptor (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
5 |
0.46 |
Binding ≤ 10μM
|
|
PE2R4-2-E |
Prostanoid EP4 Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3 |
0.48 |
Binding ≤ 10μM
|
|
PI2R-2-E |
Prostanoid IP Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
150 |
0.38 |
Binding ≤ 10μM
|
|
PE2R4-1-E |
Prostanoid EP4 Receptor (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
4 |
0.47 |
Functional ≤ 10μM
|
|
Z50587-5-O |
Homo Sapiens (cluster #5 Of 9), Other |
Other |
70 |
0.40 |
Functional ≤ 10μM |
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PE2R1_MOUSE |
P35375
|
Prostanoid EP1 Receptor, Mouse |
22 |
0.43 |
Binding ≤ 1μM
|
|
PE2R2_MOUSE |
Q62053
|
Prostanoid EP2 Receptor, Mouse |
22 |
0.43 |
Binding ≤ 1μM
|
|
PE2R3_MOUSE |
P30557
|
Prostanoid EP3 Receptor, Mouse |
5 |
0.46 |
Binding ≤ 1μM
|
|
PE2R4_MOUSE |
P32240
|
Prostanoid EP4 Receptor, Mouse |
3.1 |
0.48 |
Binding ≤ 1μM
|
|
PI2R_HUMAN |
P43119
|
Prostanoid IP Receptor, Human |
150 |
0.38 |
Binding ≤ 1μM
|
|
PE2R1_MOUSE |
P35375
|
Prostanoid EP1 Receptor, Mouse |
22 |
0.43 |
Binding ≤ 10μM
|
|
PE2R2_MOUSE |
Q62053
|
Prostanoid EP2 Receptor, Mouse |
22 |
0.43 |
Binding ≤ 10μM
|
|
PE2R3_MOUSE |
P30557
|
Prostanoid EP3 Receptor, Mouse |
5 |
0.46 |
Binding ≤ 10μM
|
|
PE2R4_MOUSE |
P32240
|
Prostanoid EP4 Receptor, Mouse |
3.1 |
0.48 |
Binding ≤ 10μM
|
|
PI2R_HUMAN |
P43119
|
Prostanoid IP Receptor, Human |
1400 |
0.33 |
Binding ≤ 10μM
|
|
Z50587 |
Z50587
|
Homo Sapiens |
70 |
0.40 |
Functional ≤ 10μM
|
|
PE2R4_MOUSE |
P32240
|
Prostanoid EP4 Receptor, Mouse |
2.5 |
0.48 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.36 |
7.89 |
-49.67 |
2 |
5 |
-1 |
98 |
353.479 |
13 |
↓
|
|
Lo
Low (pH 4.5-6)
|
3.36 |
5.91 |
-10.15 |
3 |
5 |
0 |
95 |
354.487 |
13 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.09 |
2.56 |
-70.92 |
3 |
3 |
0 |
68 |
131.175 |
5 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.09 |
0.58 |
-45.33 |
4 |
3 |
1 |
65 |
132.183 |
5 |
↓
|
|
|
|
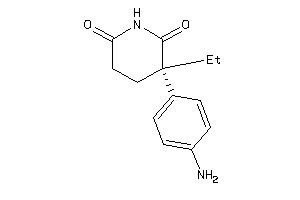
Analogs
-
1530855
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP19A-1-E |
Cytochrome P450 19A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6200 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.76 |
2.19 |
-10.22 |
3 |
4 |
0 |
72 |
232.283 |
2 |
↓
|
|
|
|
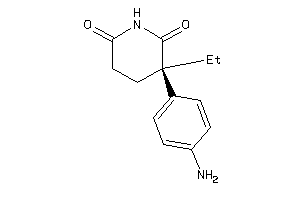
Analogs
-
1530856
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CP19A-1-E |
Cytochrome P450 19A1 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6200 |
0.43 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.76 |
2.15 |
-10.17 |
3 |
4 |
0 |
72 |
232.283 |
2 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNH2-2-E |
HERG (cluster #2 Of 5), Eukaryotic |
Eukaryotes |
210 |
0.33 |
Binding ≤ 10μM
|
|
S22A1-1-E |
Solute Carrier Family 22 Member 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
5000 |
0.27 |
Binding ≤ 10μM
|
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
9940 |
0.25 |
Binding ≤ 10μM
|
|
TOP2B-1-E |
DNA Topoisomerase II Beta (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
720 |
0.31 |
Binding ≤ 10μM |
|
TOP1-1-E |
DNA Topoisomerase I (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.38 |
Functional ≤ 10μM
|
|
TOP2A-1-E |
DNA Topoisomerase II Alpha (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.38 |
Functional ≤ 10μM
|
|
TOP2B-1-E |
DNA Topoisomerase II Beta (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
20 |
0.38 |
Functional ≤ 10μM
|
|
ADO-1-E |
Aldehyde Oxidase (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
6400 |
0.26 |
ADME/T ≤ 10μM
|
|
CP2D6-2-E |
Cytochrome P450 2D6 (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
7500 |
0.26 |
ADME/T ≤ 10μM
|
|
Z103205-3-O |
A431 (cluster #3 Of 4), Other |
Other |
5890 |
0.26 |
Functional ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
600 |
0.31 |
Functional ≤ 10μM
|
|
Z50460-4-O |
Leishmania Major (cluster #4 Of 4), Other |
Other |
1500 |
0.29 |
Functional ≤ 10μM
|
|
Z80064-8-O |
CCRF-CEM (T-cell Leukemia) (cluster #8 Of 9), Other |
Other |
139 |
0.34 |
Functional ≤ 10μM
|
|
Z80084-1-O |
CEM-VLB (Vinblastine-resistant T-cell Leukemia Cells) (cluster #1 Of 2), Other |
Other |
520 |
0.31 |
Functional ≤ 10μM
|
|
Z80085-1-O |
CEM-VM1 (Etoposide-resistant T-cell Leukemia Cells) (cluster #1 Of 2), Other |
Other |
1910 |
0.29 |
Functional ≤ 10μM
|
|
Z80089-1-O |
CHO-AA8 (cluster #1 Of 2), Other |
Other |
1 |
0.45 |
Functional ≤ 10μM
|
|
Z80115-1-O |
DC3F (cluster #1 Of 1), Other |
Other |
4 |
0.42 |
Functional ≤ 10μM
|
|
Z80117-2-O |
DC3F/AD-II (cluster #2 Of 2), Other |
Other |
47 |
0.37 |
Functional ≤ 10μM
|
|
Z80152-1-O |
HCT-8 (Ileocecal Adenocarcinoma) (cluster #1 Of 2), Other |
Other |
120 |
0.35 |
Functional ≤ 10μM
|
|
Z80156-1-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #1 Of 12), Other |
Other |
6 |
0.41 |
Functional ≤ 10μM
|
|
Z80166-1-O |
HT-29 (Colon Adenocarcinoma Cells) (cluster #1 Of 12), Other |
Other |
559 |
0.31 |
Functional ≤ 10μM |
|
Z80186-6-O |
K562 (Erythroleukemia Cells) (cluster #6 Of 11), Other |
Other |
23 |
0.38 |
Functional ≤ 10μM
|
|
Z80193-2-O |
L1210 (Lymphocytic Leukemia Cells) (cluster #2 Of 12), Other |
Other |
50 |
0.37 |
Functional ≤ 10μM
|
|
Z80224-5-O |
MCF7 (Breast Carcinoma Cells) (cluster #5 Of 14), Other |
Other |
75 |
0.36 |
Functional ≤ 10μM
|
|
Z80227-2-O |
MCF7-ADR (Breast Carcinoma Cells) (cluster #2 Of 2), Other |
Other |
1165 |
0.30 |
Functional ≤ 10μM
|
|
Z80244-7-O |
MDA-MB-468 (Breast Adenocarcinoma) (cluster #7 Of 7), Other |
Other |
490 |
0.32 |
Functional ≤ 10μM
|
|
Z80322-1-O |
NCI-H358 (Lung Carcinama Cells) (cluster #1 Of 2), Other |
Other |
150 |
0.34 |
Functional ≤ 10μM
|
|
Z80362-1-O |
P388 (Lymphoma Cells) (cluster #1 Of 8), Other |
Other |
890 |
0.30 |
Functional ≤ 10μM
|
|
Z80475-3-O |
SK-BR-3 (Breast Adenocarcinoma) (cluster #3 Of 3), Other |
Other |
60 |
0.36 |
Functional ≤ 10μM
|
|
Z80601-1-O |
XRS6 (cluster #1 Of 1), Other |
Other |
240 |
0.33 |
Functional ≤ 10μM
|
|
Z80608-4-O |
ZR-75-1 (Breast Carcinoma Cells) (cluster #4 Of 4), Other |
Other |
180 |
0.34 |
Functional ≤ 10μM
|
|
Z80612-1-O |
2008 (Ovarian Carcinoma Cells) (cluster #1 Of 2), Other |
Other |
4310 |
0.27 |
Functional ≤ 10μM
|
|
Z80682-1-O |
A549 (Lung Carcinoma Cells) (cluster #1 Of 11), Other |
Other |
3520 |
0.27 |
Functional ≤ 10μM
|
|
Z80712-7-O |
T47D (Breast Carcinoma Cells) (cluster #7 Of 7), Other |
Other |
220 |
0.33 |
Functional ≤ 10μM
|
|
Z80819-1-O |
J774.2 (cluster #1 Of 1), Other |
Other |
1485 |
0.29 |
Functional ≤ 10μM |
|
Z80846-2-O |
F460pv8/eto Cell Line (cluster #2 Of 2), Other |
Other |
40 |
0.37 |
Functional ≤ 10μM
|
|
Z80889-2-O |
NCI-H322M (Non-small Cell Lung Carcinoma Cells) (cluster #2 Of 3), Other |
Other |
210 |
0.33 |
Functional ≤ 10μM
|
|
Z80928-5-O |
HCT-116 (Colon Carcinoma Cells) (cluster #5 Of 9), Other |
Other |
700 |
0.31 |
Functional ≤ 10μM
|
|
Z81024-8-O |
NCI-H460 (Non-small Cell Lung Carcinoma) (cluster #8 Of 8), Other |
Other |
60 |
0.36 |
Functional ≤ 10μM
|
|
Z81034-6-O |
A2780 (Ovarian Carcinoma Cells) (cluster #6 Of 10), Other |
Other |
100 |
0.35 |
Functional ≤ 10μM
|
|
Z81072-6-O |
Jurkat (Acute Leukemic T-cells) (cluster #6 Of 10), Other |
Other |
80 |
0.35 |
Functional ≤ 10μM
|
|
Z81160-1-O |
Lewis Lung Carcinoma Cell Line (cluster #1 Of 1), Other |
Other |
12 |
0.40 |
Functional ≤ 10μM
|
|
Z81164-1-O |
LL Cell Line (cluster #1 Of 2), Other |
Other |
12 |
0.40 |
Functional ≤ 10μM
|
|
Z81167-1-O |
LLTC Cell Line (cluster #1 Of 1), Other |
Other |
12 |
0.40 |
Functional ≤ 10μM
|
|
Z81244-1-O |
J774 (Macrophage Cells) (cluster #1 Of 1), Other |
Other |
901 |
0.30 |
Functional ≤ 10μM |
|
Z81247-2-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #2 Of 9), Other |
Other |
9500 |
0.25 |
Functional ≤ 10μM
|
|
Z81252-10-O |
MDA-MB-231 (Breast Adenocarcinoma Cells) (cluster #10 Of 11), Other |
Other |
140 |
0.34 |
Functional ≤ 10μM
|
|
Z81280-1-O |
NCI-H226 (Non-small Cell Lung Carcinoma Cells) (cluster #1 Of 3), Other |
Other |
1010 |
0.30 |
Functional ≤ 10μM
|
|
Z81284-2-O |
NCI-H647 (cluster #2 Of 2), Other |
Other |
70 |
0.36 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
TOP2A_HUMAN |
P11388
|
DNA Topoisomerase II Alpha, Human |
1000 |
0.30 |
Binding ≤ 1μM
|
|
TOP2B_HUMAN |
Q02880
|
DNA Topoisomerase II Beta, Human |
1000 |
0.30 |
Binding ≤ 1μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
208.929613 |
0.33 |
Binding ≤ 1μM
|
|
TOP2A_HUMAN |
P11388
|
DNA Topoisomerase II Alpha, Human |
1000 |
0.30 |
Binding ≤ 10μM
|
|
TOP2B_HUMAN |
Q02880
|
DNA Topoisomerase II Beta, Human |
1000 |
0.30 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
208.929613 |
0.33 |
Binding ≤ 10μM
|
|
S22A1_HUMAN |
O15245
|
Solute Carrier Family 22 Member 1, Human |
5000 |
0.27 |
Binding ≤ 10μM
|
|
Z80612 |
Z80612
|
2008 (Ovarian Carcinoma Cells) |
4310 |
0.27 |
Functional ≤ 10μM
|
|
Z81034 |
Z81034
|
A2780 (Ovarian Carcinoma Cells) |
100 |
0.35 |
Functional ≤ 10μM
|
|
Z103205 |
Z103205
|
A431 |
170 |
0.34 |
Functional ≤ 10μM
|
|
Z80682 |
Z80682
|
A549 (Lung Carcinoma Cells) |
30 |
0.38 |
Functional ≤ 10μM
|
|
Z80064 |
Z80064
|
CCRF-CEM (T-cell Leukemia) |
130 |
0.34 |
Functional ≤ 10μM
|
|
Z80084 |
Z80084
|
CEM-VLB (Vinblastine-resistant T-cell Leukemia Cells) |
520 |
0.31 |
Functional ≤ 10μM
|
|
Z80085 |
Z80085
|
CEM-VM1 (Etoposide-resistant T-cell Leukemia Cells) |
1910 |
0.29 |
Functional ≤ 10μM
|
|
Z80089 |
Z80089
|
CHO-AA8 |
0.6 |
0.46 |
Functional ≤ 10μM
|
|
Z80115 |
Z80115
|
DC3F |
4 |
0.42 |
Functional ≤ 10μM
|
|
Z80117 |
Z80117
|
DC3F/AD-II |
47 |
0.37 |
Functional ≤ 10μM
|
|
TOP1_MOUSE |
Q04750
|
DNA Topoisomerase I, Mouse |
12 |
0.40 |
Functional ≤ 10μM
|
|
TOP1_HUMAN |
P11387
|
DNA Topoisomerase I, Human |
37 |
0.37 |
Functional ≤ 10μM
|
|
TOP2A_MOUSE |
Q01320
|
DNA Topoisomerase II Alpha, Mouse |
12 |
0.40 |
Functional ≤ 10μM
|
|
TOP2A_HUMAN |
P11388
|
DNA Topoisomerase II Alpha, Human |
37 |
0.37 |
Functional ≤ 10μM
|
|
TOP2B_MOUSE |
Q64511
|
DNA Topoisomerase II Beta, Mouse |
12 |
0.40 |
Functional ≤ 10μM
|
|
TOP2B_HUMAN |
Q02880
|
DNA Topoisomerase II Beta, Human |
37 |
0.37 |
Functional ≤ 10μM
|
|
Z80846 |
Z80846
|
F460pv8/eto Cell Line |
40 |
0.37 |
Functional ≤ 10μM
|
|
Z80928 |
Z80928
|
HCT-116 (Colon Carcinoma Cells) |
6300 |
0.26 |
Functional ≤ 10μM
|
|
Z80152 |
Z80152
|
HCT-8 (Ileocecal Adenocarcinoma) |
120 |
0.35 |
Functional ≤ 10μM
|
|
Z81247 |
Z81247
|
HeLa (Cervical Adenocarcinoma Cells) |
12 |
0.40 |
Functional ≤ 10μM
|
|
Z80156 |
Z80156
|
HL-60 (Promyeloblast Leukemia Cells) |
1150 |
0.30 |
Functional ≤ 10μM
|
|
Z80166 |
Z80166
|
HT-29 (Colon Adenocarcinoma Cells) |
559 |
0.31 |
Functional ≤ 10μM
|
|
Z81244 |
Z81244
|
J774 (Macrophage Cells) |
900.6 |
0.30 |
Functional ≤ 10μM
|
|
Z80819 |
Z80819
|
J774.2 |
1353.9 |
0.29 |
Functional ≤ 10μM
|
|
Z81072 |
Z81072
|
Jurkat (Acute Leukemic T-cells) |
1000 |
0.30 |
Functional ≤ 10μM
|
|
Z80186 |
Z80186
|
K562 (Erythroleukemia Cells) |
23 |
0.38 |
Functional ≤ 10μM
|
|
Z80193 |
Z80193
|
L1210 (Lymphocytic Leukemia Cells) |
33 |
0.37 |
Functional ≤ 10μM
|
|
Z50460 |
Z50460
|
Leishmania Major |
1500 |
0.29 |
Functional ≤ 10μM
|
|
Z81160 |
Z81160
|
Lewis Lung Carcinoma Cell Line |
12 |
0.40 |
Functional ≤ 10μM
|
|
Z81164 |
Z81164
|
LL Cell Line |
12 |
0.40 |
Functional ≤ 10μM
|
|
Z81167 |
Z81167
|
LLTC Cell Line |
12 |
0.40 |
Functional ≤ 10μM
|
|
Z80224 |
Z80224
|
MCF7 (Breast Carcinoma Cells) |
5130 |
0.26 |
Functional ≤ 10μM
|
|
Z80227 |
Z80227
|
MCF7-ADR (Breast Carcinoma Cells) |
1165 |
0.30 |
Functional ≤ 10μM
|
|
Z81252 |
Z81252
|
MDA-MB-231 (Breast Adenocarcinoma Cells) |
140 |
0.34 |
Functional ≤ 10μM
|
|
Z80244 |
Z80244
|
MDA-MB-468 (Breast Adenocarcinoma) |
490 |
0.32 |
Functional ≤ 10μM
|
|
Z81280 |
Z81280
|
NCI-H226 (Non-small Cell Lung Carcinoma Cells) |
1010 |
0.30 |
Functional ≤ 10μM
|
|
Z80889 |
Z80889
|
NCI-H322M (Non-small Cell Lung Carcinoma Cells) |
210 |
0.33 |
Functional ≤ 10μM
|
|
Z80322 |
Z80322
|
NCI-H358 (Lung Carcinama Cells) |
150 |
0.34 |
Functional ≤ 10μM
|
|
Z81024 |
Z81024
|
NCI-H460 (Non-small Cell Lung Carcinoma) |
200 |
0.33 |
Functional ≤ 10μM
|
|
Z81284 |
Z81284
|
NCI-H647 |
70 |
0.36 |
Functional ≤ 10μM
|
|
Z80362 |
Z80362
|
P388 (Lymphoma Cells) |
12 |
0.40 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
1000 |
0.30 |
Functional ≤ 10μM
|
|
Z80475 |
Z80475
|
SK-BR-3 (Breast Adenocarcinoma) |
60 |
0.36 |
Functional ≤ 10μM
|
|
Z80712 |
Z80712
|
T47D (Breast Carcinoma Cells) |
220 |
0.33 |
Functional ≤ 10μM
|
|
Z80601 |
Z80601
|
XRS6 |
240 |
0.33 |
Functional ≤ 10μM
|
|
Z80608 |
Z80608
|
ZR-75-1 (Breast Carcinoma Cells) |
180 |
0.34 |
Functional ≤ 10μM
|
|
ADO_RABIT |
P80456
|
Aldehyde Oxidase, Rabit |
60 |
0.36 |
ADME/T ≤ 10μM
|
|
ADO_RAT |
Q9Z0U5
|
Aldehyde Oxidase, Rat |
3000 |
0.28 |
ADME/T ≤ 10μM
|
|
ADO_HUMAN |
Q06278
|
Aldehyde Oxidase, Human |
3200 |
0.27 |
ADME/T ≤ 10μM
|
|
CP2D6_HUMAN |
P10635
|
Cytochrome P450 2D6, Human |
7500 |
0.26 |
ADME/T ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.91 |
6.33 |
-44.52 |
2 |
6 |
0 |
84 |
393.468 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.91 |
5.83 |
-16.62 |
2 |
6 |
0 |
80 |
393.468 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.91 |
5.94 |
-47.33 |
1 |
6 |
-1 |
82 |
392.46 |
5 |
↓
|
|
|
|
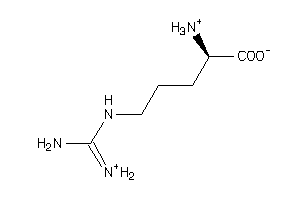
Analogs
-
1532749
-
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS2-7-E |
Nitric Oxide Synthase, Inducible (cluster #7 Of 9), Eukaryotic |
Eukaryotes |
7000 |
0.60 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
NOS2_HUMAN |
P35228
|
Nitric Oxide Synthase, Inducible, Human |
7000 |
0.60 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.63 |
0.33 |
-74.97 |
8 |
6 |
1 |
131 |
175.212 |
6 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.64 |
10.11 |
-7.64 |
0 |
2 |
0 |
20 |
328.209 |
1 |
↓
|
|
|
|
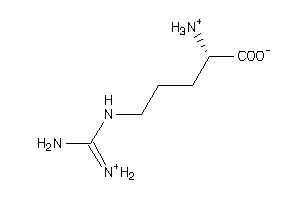
Analogs
-
1532525
-
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-3.63 |
0.35 |
-74.53 |
8 |
6 |
1 |
131 |
175.212 |
6 |
↓
|
|
|
|
Analogs
-
1846151
-
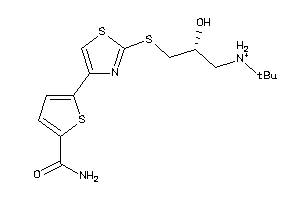
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.43 |
1.85 |
-51.09 |
5 |
5 |
1 |
93 |
372.561 |
8 |
↓
|
|
|
|
Analogs
-
1542905
-
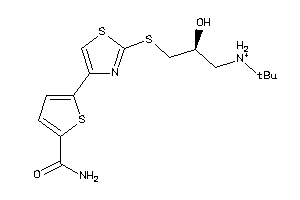
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.43 |
-7.63 |
-51.16 |
5 |
5 |
1 |
92 |
372.561 |
8 |
↓
|
|
|
|
Analogs
-
113415
-
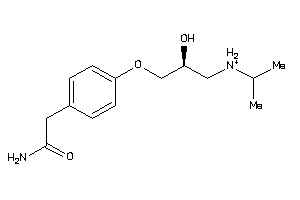
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA3R-3-E |
Adenosine Receptor A3 (cluster #3 Of 6), Eukaryotic |
Eukaryotes |
2 |
0.64 |
Binding ≤ 10μM
|
|
ADRB1-2-E |
Beta-1 Adrenergic Receptor (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
600 |
0.46 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.72 |
0.6 |
-46.26 |
5 |
5 |
1 |
89 |
267.349 |
8 |
↓
|
|
Hi
High (pH 8-9.5)
|
0.72 |
-0.67 |
-12.7 |
4 |
5 |
0 |
85 |
266.341 |
8 |
↓
|
|
|
|
Analogs
-
58
-
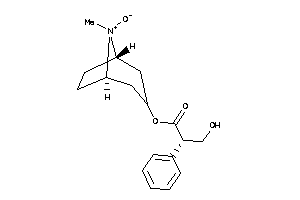
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.47 |
-0.24 |
-19.78 |
1 |
5 |
0 |
63 |
305.374 |
5 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.74 |
-0.67 |
-5.46 |
2 |
5 |
0 |
75 |
184.195 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.04 |
-3.18 |
-35.95 |
1 |
5 |
-1 |
82 |
183.187 |
2 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.52 |
1.64 |
-57.87 |
2 |
7 |
0 |
103 |
424.497 |
10 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.88 |
2.25 |
-36.49 |
1 |
2 |
1 |
13 |
290.471 |
7 |
↓
|
|
|
|
Analogs
-
70
-
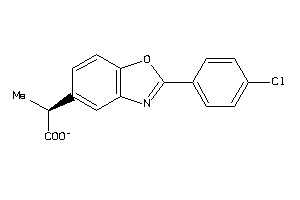
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.53 |
-0.61 |
-46.47 |
0 |
4 |
-1 |
66 |
300.721 |
3 |
↓
|
|
|
|
Analogs
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
|
|
Analogs
-
12959181
-
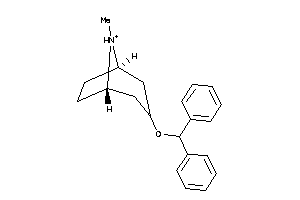
-
16969857
-
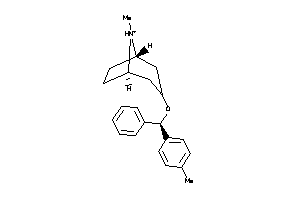
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
SC6A3_HUMAN |
Q01959
|
Dopamine Transporter, Human |
312 |
0.40 |
Binding ≤ 1μM
|
|
SC6A3_RAT |
P23977
|
Dopamine Transporter, Rat |
118 |
0.42 |
Binding ≤ 1μM
|
|
ACM1_RAT |
P08482
|
Muscarinic Acetylcholine Receptor M1, Rat |
0.59 |
0.56 |
Binding ≤ 1μM
|
|
ACM2_RAT |
P10980
|
Muscarinic Acetylcholine Receptor M2, Rat |
2.6 |
0.52 |
Binding ≤ 1μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
130 |
0.42 |
Binding ≤ 1μM
|
|
SC6A3_HUMAN |
Q01959
|
Dopamine Transporter, Human |
312 |
0.40 |
Binding ≤ 10μM
|
|
SC6A3_RAT |
P23977
|
Dopamine Transporter, Rat |
118 |
0.42 |
Binding ≤ 10μM
|
|
ACM1_RAT |
P08482
|
Muscarinic Acetylcholine Receptor M1, Rat |
0.59 |
0.56 |
Binding ≤ 10μM
|
|
ACM2_RAT |
P10980
|
Muscarinic Acetylcholine Receptor M2, Rat |
2.6 |
0.52 |
Binding ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
130 |
0.42 |
Binding ≤ 10μM
|
|
SC6A4_RAT |
P31652
|
Serotonin Transporter, Rat |
5150 |
0.32 |
Binding ≤ 10μM
|
|
SC6A3_RAT |
P23977
|
Dopamine Transporter, Rat |
403 |
0.39 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
1995.26231 |
0.35 |
Functional ≤ 10μM
|
|
Z50597 |
Z50597
|
Rattus Norvegicus |
403 |
0.39 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.27 |
12.85 |
-32.6 |
1 |
2 |
1 |
14 |
308.445 |
4 |
↓
|
|
|
|
Analogs
-
6578890
-
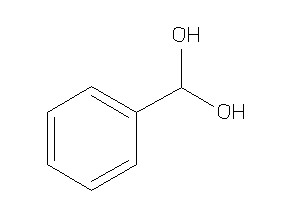
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.27 |
1.82 |
-4.35 |
1 |
1 |
0 |
20 |
108.14 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
184 |
0.35 |
Binding ≤ 1μM
|
|
SCN1A_HUMAN |
P35498
|
Sodium Channel Protein Type I Alpha Subunit, Human |
840 |
0.32 |
Binding ≤ 1μM
|
|
SCN2A_HUMAN |
Q99250
|
Sodium Channel Protein Type II Alpha Subunit, Human |
840 |
0.32 |
Binding ≤ 1μM
|
|
SCN3A_HUMAN |
Q9NY46
|
Sodium Channel Protein Type III Alpha Subunit, Human |
840 |
0.32 |
Binding ≤ 1μM
|
|
SCN8A_HUMAN |
Q9UQD0
|
Sodium Channel Protein Type VIII Alpha Subunit, Human |
840 |
0.32 |
Binding ≤ 1μM
|
|
LEF_BACAN |
P15917
|
Anthrax Lethal Factor, Bacan |
4800 |
0.28 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
184 |
0.35 |
Binding ≤ 10μM
|
|
SCN1A_HUMAN |
P35498
|
Sodium Channel Protein Type I Alpha Subunit, Human |
840 |
0.32 |
Binding ≤ 10μM
|
|
SCN2A_HUMAN |
Q99250
|
Sodium Channel Protein Type II Alpha Subunit, Human |
840 |
0.32 |
Binding ≤ 10μM
|
|
SCN3A_HUMAN |
Q9NY46
|
Sodium Channel Protein Type III Alpha Subunit, Human |
840 |
0.32 |
Binding ≤ 10μM
|
|
SCN8A_HUMAN |
Q9UQD0
|
Sodium Channel Protein Type VIII Alpha Subunit, Human |
840 |
0.32 |
Binding ≤ 10μM
|
|
KCNH2_HUMAN |
Q12809
|
HERG, Human |
550 |
0.32 |
Functional ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
2500 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.97 |
14.18 |
-37.2 |
1 |
3 |
1 |
17 |
367.557 |
10 |
↓
|
|
|
|
Analogs
-
895466
-
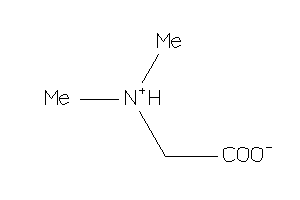
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.33 |
5.26 |
-31.5 |
0 |
3 |
0 |
40 |
117.148 |
2 |
↓
|
|
|
|
|
|
|
|
|
|
Analogs
-
3830314
-
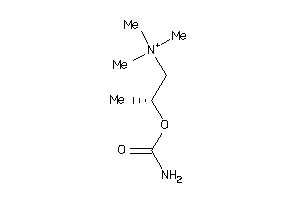
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-4.01 |
2.07 |
-34.67 |
2 |
4 |
1 |
52 |
161.225 |
4 |
↓
|
|
|
|
Analogs
-
1530570
-
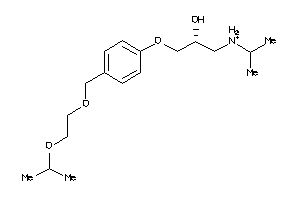
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
3162 |
0.33 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
3162.27766 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.30 |
4.82 |
-43.49 |
3 |
5 |
1 |
65 |
326.457 |
12 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.30 |
3.64 |
-8.33 |
2 |
5 |
0 |
60 |
325.449 |
12 |
↓
|
|
|
|
Analogs
-
1530569
-
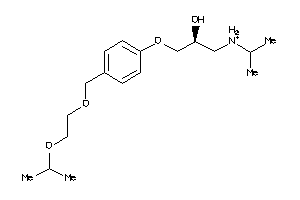
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
3162 |
0.33 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
3162.27766 |
0.33 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.30 |
4.8 |
-43.51 |
3 |
5 |
1 |
65 |
326.457 |
12 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.30 |
3.53 |
-8.45 |
2 |
5 |
0 |
60 |
325.449 |
12 |
↓
|
|
|
|
Analogs
-
1542901
-
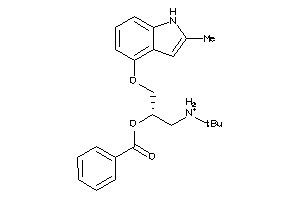
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.14 |
1.15 |
-32.83 |
3 |
5 |
1 |
67 |
381.496 |
9 |
↓
|
|
|
|
Analogs
-
2032320
-
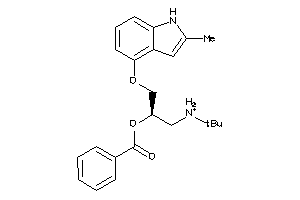
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.14 |
11.58 |
-32.31 |
3 |
5 |
1 |
68 |
381.496 |
9 |
↓
|
|
Hi
High (pH 8-9.5)
|
5.14 |
10.57 |
-9.55 |
2 |
5 |
0 |
63 |
380.488 |
9 |
↓
|
|
|
|
Analogs
-
95
-
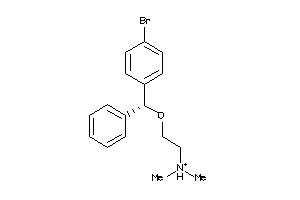
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.31 |
1.67 |
-36.2 |
1 |
2 |
1 |
13 |
335.265 |
6 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
PTAFR-1-E |
Platelet Activating Factor Receptor (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.42 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.17 |
8.92 |
-10.07 |
0 |
4 |
0 |
43 |
393.697 |
1 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.17 |
9.86 |
-33.76 |
1 |
4 |
1 |
45 |
394.705 |
1 |
↓
|
|