|
|
Analogs
-
8214760
-
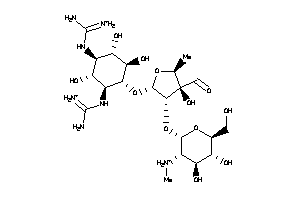
-
8143632
-
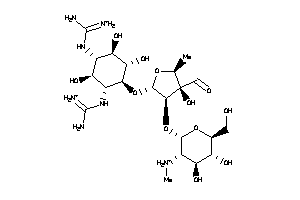
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.28 |
-14.78 |
-150.46 |
19 |
19 |
3 |
339 |
584.604 |
11 |
↓
|
|
Mid
Mid (pH 6-8)
|
-5.28 |
-16.18 |
-95.14 |
18 |
19 |
2 |
335 |
583.596 |
11 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.51 |
-26.31 |
-19.41 |
14 |
19 |
0 |
321 |
645.608 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
-5.51 |
-25.34 |
-49.38 |
15 |
19 |
1 |
326 |
646.616 |
9 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-5.51 |
-26.09 |
-18.7 |
14 |
19 |
0 |
321 |
645.608 |
9 |
↓
|
|
Mid
Mid (pH 6-8)
|
-5.51 |
-25.4 |
-47.44 |
15 |
19 |
1 |
326 |
646.616 |
9 |
↓
|
|
|
|
Analogs
-
33845547
-
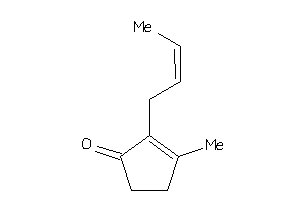
-
1531140
-
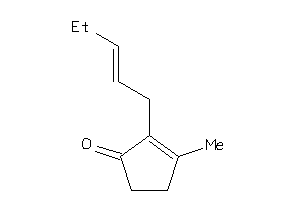
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.56 |
2.24 |
-6.83 |
0 |
1 |
0 |
17 |
164.248 |
3 |
↓
|
|
|
|
Analogs
-
35525
-
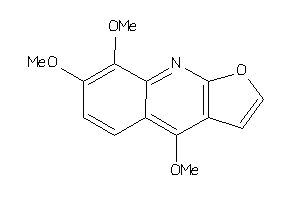
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.39 |
0.76 |
-13.88 |
1 |
5 |
0 |
65 |
245.234 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.39 |
1.69 |
-51.13 |
0 |
5 |
-1 |
68 |
244.226 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.60 |
7.87 |
-12.94 |
0 |
6 |
0 |
67 |
342.347 |
5 |
↓
|
|
|
|
Analogs
-
608165
-
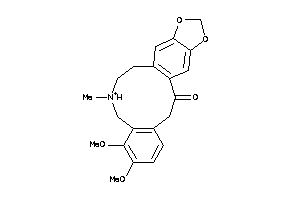
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426-6-O |
Plasmodium Falciparum (isolate K1 / Thailand) (cluster #6 Of 9), Other |
Other |
5080 |
0.27 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50426 |
Z50426
|
Plasmodium Falciparum (isolate K1 / Thailand) |
5080 |
0.27 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.70 |
7.84 |
-11.18 |
0 |
6 |
0 |
57 |
369.417 |
2 |
↓
|
|
|
|
Analogs
-
5699358
-
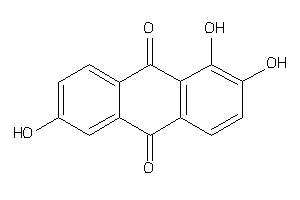
-
5699362
-
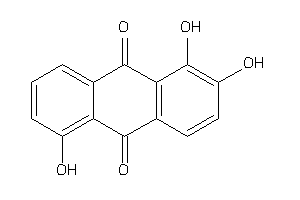
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.91 |
3.61 |
-10.65 |
2 |
4 |
0 |
75 |
240.214 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.91 |
4.53 |
-45.52 |
1 |
4 |
-1 |
77 |
239.206 |
0 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.03 |
-7.26 |
-11.03 |
5 |
7 |
0 |
113 |
158.117 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.78 |
-12.09 |
-35.58 |
6 |
7 |
1 |
122 |
159.125 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.78 |
-13.67 |
-13.57 |
5 |
7 |
0 |
120 |
158.117 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-2.03 |
-7.26 |
-11.05 |
5 |
7 |
0 |
113 |
158.117 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.78 |
-13.65 |
-13.6 |
5 |
7 |
0 |
120 |
158.117 |
1 |
↓
|
|
Hi
High (pH 8-9.5)
|
-2.78 |
-12.06 |
-35.62 |
6 |
7 |
1 |
122 |
159.125 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
4.74 |
4.12 |
-16.76 |
4 |
6 |
0 |
115 |
376.364 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
4.74 |
5.13 |
-66.02 |
3 |
6 |
-1 |
118 |
375.356 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.56 |
7.4 |
-123 |
1 |
6 |
-2 |
118 |
374.348 |
0 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
-
370772
-
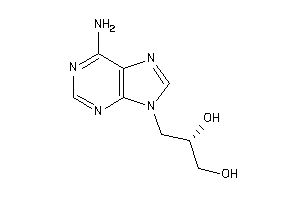
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.97 |
-1.19 |
-16.13 |
4 |
7 |
0 |
110 |
209.209 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.97 |
-0.93 |
-38.71 |
5 |
7 |
1 |
111 |
210.217 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.97 |
-5.3 |
-37.36 |
5 |
7 |
1 |
111 |
210.217 |
3 |
↓
|
|
|
|
Analogs
-
370776
-
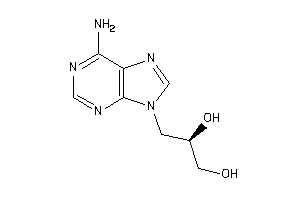
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AA2AR-1-E |
Adenosine A2a Receptor (cluster #1 Of 4), Eukaryotic |
Eukaryotes |
8500 |
0.47 |
Binding ≤ 10μM
|
|
SAHH-1-E |
Adenosylhomocysteinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
280 |
0.61 |
Binding ≤ 10μM |
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-0.97 |
-1.27 |
-15 |
4 |
7 |
0 |
110 |
209.209 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.97 |
-5.39 |
-36.69 |
5 |
7 |
1 |
111 |
210.217 |
3 |
↓
|
|
Lo
Low (pH 4.5-6)
|
-0.97 |
-0.9 |
-38.73 |
5 |
7 |
1 |
111 |
210.217 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.54 |
7.24 |
-14.5 |
0 |
4 |
0 |
57 |
204.181 |
2 |
↓
|
|
|
|
Analogs
-
4015531
-
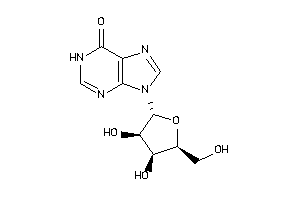
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80583-7-O |
Vero (Kidney Cells) (cluster #7 Of 7), Other |
Other |
0 |
0.00 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.82 |
-4.43 |
-23.87 |
4 |
9 |
0 |
133 |
268.229 |
2 |
↓
|
|
Hi
High (pH 8-9.5)
|
-1.36 |
-6.52 |
-59.54 |
3 |
9 |
-1 |
137 |
267.221 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80156-12-O |
HL-60 (Promyeloblast Leukemia Cells) (cluster #12 Of 12), Other |
Other |
9500 |
0.50 |
Functional ≤ 10μM
|
|
Z80211-5-O |
LoVo (Colon Adenocarcinoma Cells) (cluster #5 Of 5), Other |
Other |
7300 |
0.51 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.33 |
3.16 |
-12.54 |
1 |
4 |
0 |
60 |
192.17 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z101794-1-O |
Felid Herpesvirus 1 (cluster #1 Of 1), Other |
Other |
1000 |
0.53 |
Functional ≤ 10μM |
|
Z50515-2-O |
Human Herpesvirus 2 (cluster #2 Of 2), Other |
Other |
9600 |
0.44 |
Functional ≤ 10μM
|
|
Z50517-1-O |
Human Herpesvirus 5 Strain AD169 (cluster #1 Of 2), Other |
Other |
7600 |
0.45 |
Functional ≤ 10μM
|
|
Z50518-3-O |
Human Herpesvirus 4 (cluster #3 Of 5), Other |
Other |
6900 |
0.45 |
Functional ≤ 10μM
|
|
Z50527-1-O |
Human Herpesvirus 3 (cluster #1 Of 3), Other |
Other |
900 |
0.53 |
Functional ≤ 10μM
|
|
Z50527-3-O |
Human Herpesvirus 3 (cluster #3 Of 3), Other |
Other |
4900 |
0.46 |
Functional ≤ 10μM |
|
Z50530-1-O |
Human Herpesvirus 5 (cluster #1 Of 5), Other |
Other |
7500 |
0.45 |
Functional ≤ 10μM
|
|
Z50587-1-O |
Homo Sapiens (cluster #1 Of 9), Other |
Other |
90 |
0.62 |
Functional ≤ 10μM
|
|
Z50600-1-O |
Vaccinia Virus (cluster #1 Of 2), Other |
Other |
384 |
0.56 |
Functional ≤ 10μM
|
|
Z50602-2-O |
Human Herpesvirus 1 (cluster #2 Of 5), Other |
Other |
990 |
0.53 |
Functional ≤ 10μM
|
|
Z50606-1-O |
Hepatitis B Virus (cluster #1 Of 3), Other |
Other |
20 |
0.67 |
Functional ≤ 10μM
|
|
Z80040-1-O |
BHK-21 (Kidney Cells) (cluster #1 Of 1), Other |
Other |
2000 |
0.50 |
Functional ≤ 10μM
|
|
Z80291-2-O |
MRC5 (Embryonic Lung Fibroblast Cells) (cluster #2 Of 3), Other |
Other |
5000 |
0.46 |
Functional ≤ 10μM
|
|
Z80414-1-O |
Raji (B-lymphoblastic Cells) (cluster #1 Of 3), Other |
Other |
2900 |
0.48 |
Functional ≤ 10μM
|
|
Z80583-7-O |
Vero (Kidney Cells) (cluster #7 Of 7), Other |
Other |
7540 |
0.45 |
Functional ≤ 10μM
|
|
Z80942-1-O |
HEL (Embryonic Lung Cells) (cluster #1 Of 2), Other |
Other |
900 |
0.53 |
Functional ≤ 10μM
|
|
Z80954-1-O |
HFF (Foreskin Fibroblasts) (cluster #1 Of 4), Other |
Other |
8100 |
0.45 |
Functional ≤ 10μM
|
|
Z81247-5-O |
HeLa (Cervical Adenocarcinoma Cells) (cluster #5 Of 9), Other |
Other |
8710 |
0.44 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
-1.61 |
-0.6 |
-19.69 |
4 |
8 |
0 |
119 |
225.208 |
4 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.96 |
2.54 |
-7.62 |
2 |
3 |
0 |
37 |
202.257 |
1 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.50 |
7.43 |
-13 |
0 |
6 |
0 |
57 |
369.417 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.48 |
2.35 |
-11.66 |
1 |
4 |
0 |
55 |
202.213 |
0 |
↓
|
|
Ref
Reference (pH 7)
|
0.48 |
2.35 |
-11.88 |
1 |
4 |
0 |
55 |
202.213 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.75 |
4.95 |
-40.62 |
0 |
3 |
-1 |
57 |
187.174 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.71 |
4.76 |
-9.6 |
0 |
3 |
0 |
51 |
188.182 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
1.75 |
3.99 |
-26.74 |
1 |
3 |
0 |
54 |
188.182 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH9-11-E |
Carbonic Anhydrase IX (cluster #11 Of 11), Eukaryotic |
Eukaryotes |
482 |
0.74 |
Binding ≤ 10μM
|
|
Q43570-1-E |
Zn Finger Protein (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
754 |
0.71 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.51 |
3.16 |
-12.7 |
1 |
3 |
0 |
50 |
162.144 |
0 |
↓
|
|
|
|
|
|
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.88 |
2.78 |
-13.15 |
0 |
4 |
0 |
45 |
229.235 |
2 |
↓
|
|
Mid
Mid (pH 6-8)
|
2.88 |
3.84 |
-22.37 |
1 |
4 |
1 |
46 |
230.243 |
2 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.98 |
6.17 |
-15.92 |
2 |
5 |
0 |
70 |
203.249 |
2 |
↓
|
|
|
|
Analogs
-
1583701
-
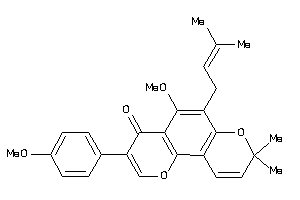
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
5.73 |
9.29 |
-12.67 |
2 |
5 |
0 |
80 |
404.462 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.55 |
7.89 |
-10.72 |
0 |
4 |
0 |
53 |
258.273 |
2 |
↓
|
|
|
|
Analogs
-
6117388
-
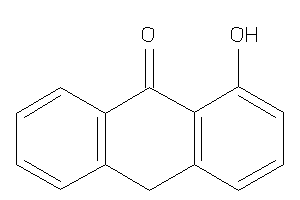
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.13 |
3.69 |
-11.62 |
2 |
4 |
0 |
75 |
240.214 |
0 |
↓
|
|
|
|
Analogs
-
34583707
-
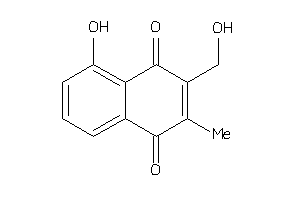
-
39283830
-
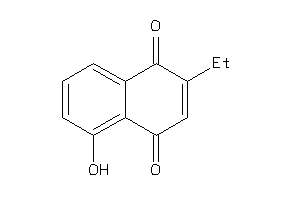
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z50425-11-O |
Plasmodium Falciparum (cluster #11 Of 22), Other |
Other |
270 |
0.66 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.78 |
4.24 |
-7.85 |
1 |
3 |
0 |
54 |
188.182 |
0 |
↓
|
|
|
|
Analogs
-
622126
-
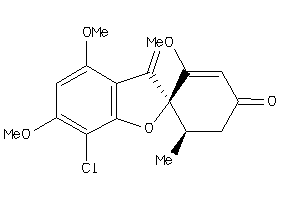
-
537809
-
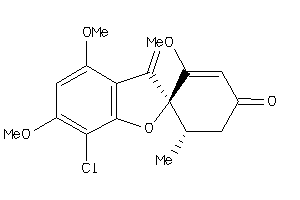
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80874-1-O |
CEM (T-cell Leukemia) (cluster #1 Of 7), Other |
Other |
10000 |
0.29 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
Z80874 |
Z80874
|
CEM (T-cell Leukemia) |
10000 |
0.29 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.58 |
4.81 |
-12.73 |
0 |
6 |
0 |
71 |
352.77 |
3 |
↓
|
|
|
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH1-1-E |
Carbonic Anhydrase I (cluster #1 Of 12), Eukaryotic |
Eukaryotes |
2420 |
0.37 |
Binding ≤ 10μM
|
|
CAH12-2-E |
Carbonic Anhydrase XII (cluster #2 Of 9), Eukaryotic |
Eukaryotes |
4720 |
0.36 |
Binding ≤ 10μM
|
|
CAH15-4-E |
Carbonic Anhydrase 15 (cluster #4 Of 6), Eukaryotic |
Eukaryotes |
7680 |
0.34 |
Binding ≤ 10μM
|
|
CAH2-1-E |
Carbonic Anhydrase II (cluster #1 Of 15), Eukaryotic |
Eukaryotes |
1840 |
0.38 |
Binding ≤ 10μM
|
|
CAH3-2-E |
Carbonic Anhydrase III (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
3580 |
0.36 |
Binding ≤ 10μM
|
|
CAH4-3-E |
Carbonic Anhydrase IV (cluster #3 Of 16), Eukaryotic |
Eukaryotes |
4900 |
0.35 |
Binding ≤ 10μM
|
|
CAH5A-6-E |
Carbonic Anhydrase VA (cluster #6 Of 10), Eukaryotic |
Eukaryotes |
4210 |
0.36 |
Binding ≤ 10μM
|
|
CAH5B-4-E |
Carbonic Anhydrase VB (cluster #4 Of 9), Eukaryotic |
Eukaryotes |
4020 |
0.36 |
Binding ≤ 10μM
|
|
CAH6-1-E |
Carbonic Anhydrase VI (cluster #1 Of 8), Eukaryotic |
Eukaryotes |
4910 |
0.35 |
Binding ≤ 10μM
|
|
CAH7-2-E |
Carbonic Anhydrase VII (cluster #2 Of 8), Eukaryotic |
Eukaryotes |
450 |
0.42 |
Binding ≤ 10μM
|
|
CAH9-3-E |
Carbonic Anhydrase IX (cluster #3 Of 11), Eukaryotic |
Eukaryotes |
5030 |
0.35 |
Binding ≤ 10μM
|
|
PGH1-2-E |
Cyclooxygenase-1 (cluster #2 Of 6), Eukaryotic |
Eukaryotes |
2000 |
0.38 |
Binding ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
450 |
0.42 |
Binding ≤ 1μM
|
|
CAH15_MOUSE |
Q99N23
|
Carbonic Anhydrase 15, Mouse |
7680 |
0.34 |
Binding ≤ 10μM
|
|
CAH1_HUMAN |
P00915
|
Carbonic Anhydrase I, Human |
2420 |
0.37 |
Binding ≤ 10μM
|
|
CAH2_HUMAN |
P00918
|
Carbonic Anhydrase II, Human |
1840 |
0.38 |
Binding ≤ 10μM
|
|
CAH3_HUMAN |
P07451
|
Carbonic Anhydrase III, Human |
3580 |
0.36 |
Binding ≤ 10μM
|
|
CAH4_HUMAN |
P22748
|
Carbonic Anhydrase IV, Human |
4900 |
0.35 |
Binding ≤ 10μM
|
|
CAH9_HUMAN |
Q16790
|
Carbonic Anhydrase IX, Human |
5030 |
0.35 |
Binding ≤ 10μM
|
|
CAH5A_HUMAN |
P35218
|
Carbonic Anhydrase VA, Human |
4210 |
0.36 |
Binding ≤ 10μM
|
|
CAH5B_HUMAN |
Q9Y2D0
|
Carbonic Anhydrase VB, Human |
4020 |
0.36 |
Binding ≤ 10μM
|
|
CAH6_HUMAN |
P23280
|
Carbonic Anhydrase VI, Human |
4910 |
0.35 |
Binding ≤ 10μM
|
|
CAH7_HUMAN |
P43166
|
Carbonic Anhydrase VII, Human |
450 |
0.42 |
Binding ≤ 10μM
|
|
CAH12_HUMAN |
O43570
|
Carbonic Anhydrase XII, Human |
4720 |
0.36 |
Binding ≤ 10μM
|
|
PGH1_SHEEP |
P05979
|
Cyclooxygenase-1, Sheep |
2000 |
0.38 |
Binding ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
1.37 |
-4.31 |
-13.19 |
5 |
6 |
0 |
110 |
290.271 |
1 |
↓
|
|
|
|
Analogs
-
4098705
-
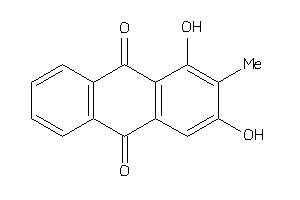
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.01 |
1.74 |
-10.81 |
3 |
5 |
0 |
95 |
270.24 |
0 |
↓
|
|
Hi
High (pH 8-9.5)
|
3.01 |
2.52 |
-45.75 |
2 |
5 |
-1 |
98 |
269.232 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.02 |
3.13 |
-21.32 |
0 |
4 |
0 |
55 |
164.164 |
1 |
↓
|
|
|
|
|
|
|
Analogs
-
38778360
-
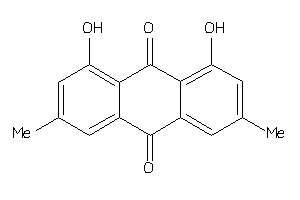
-
39284619
-
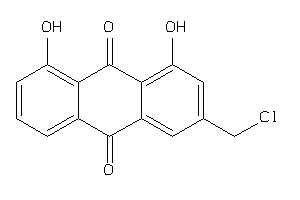
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.54 |
4.57 |
-10.59 |
2 |
4 |
0 |
75 |
254.241 |
0 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ATP4A-1-E |
Potassium-transporting ATPase Alpha Chain 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
501 |
0.37 |
Binding ≤ 10μM
|
|
ATP4B-1-E |
Potassium-transporting ATPase Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
501 |
0.37 |
Binding ≤ 10μM
|
|
ATP4A-1-E |
Potassium-transporting ATPase Alpha Chain 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
370 |
0.38 |
Functional ≤ 10μM
|
|
ATP4B-1-E |
Potassium-transporting ATPase Beta Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
370 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
3.99 |
-43.94 |
2 |
6 |
1 |
78 |
346.432 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.41 |
3.56 |
-24.45 |
1 |
6 |
0 |
77 |
345.424 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
ATP4A-1-E |
Potassium-transporting ATPase Alpha Chain 1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
501 |
0.37 |
Binding ≤ 10μM
|
|
ATP4B-1-E |
Potassium-transporting ATPase Beta Chain (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
501 |
0.37 |
Binding ≤ 10μM
|
|
ATP4A-1-E |
Potassium-transporting ATPase Alpha Chain 1 (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
370 |
0.38 |
Functional ≤ 10μM
|
|
ATP4B-1-E |
Potassium-transporting ATPase Beta Chain (cluster #1 Of 2), Eukaryotic |
Eukaryotes |
370 |
0.38 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.41 |
4.06 |
-43.78 |
2 |
6 |
1 |
78 |
346.432 |
5 |
↓
|
|
Hi
High (pH 8-9.5)
|
2.41 |
3.62 |
-13.05 |
1 |
6 |
0 |
77 |
345.424 |
5 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Clustered Target Annotations
| Code |
Organism Class |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AAK1-2-E |
Adaptor-associated Kinase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
35 |
0.61 |
Binding ≤ 10μM
|
|
AURKA-2-E |
Serine/threonine-protein Kinase Aurora-A (cluster #2 Of 3), Eukaryotic |
Eukaryotes |
980 |
0.49 |
Binding ≤ 10μM
|
|
AURKC-2-E |
Serine/threonine-protein Kinase Aurora-C (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
5300 |
0.43 |
Binding ≤ 10μM
|
|
BMP2K-2-E |
BMP-2-inducible Protein Kinase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
25 |
0.63 |
Binding ≤ 10μM
|
|
CDK16-1-E |
Serine/threonine-protein Kinase PCTAIRE-1 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
180 |
0.56 |
Binding ≤ 10μM
|
|
CDK2-5-E |
Cyclin-dependent Kinase 2 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
3200 |
0.45 |
Binding ≤ 10μM
|
|
CLK2-2-E |
Dual Specificity Protein Kinase CLK2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
300 |
0.54 |
Binding ≤ 10μM
|
|
CLK4-2-E |
Dual Specificity Protein Kinase CLK4 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
260 |
0.54 |
Binding ≤ 10μM
|
|
DAPK2-2-E |
Death-associated Protein Kinase 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
610 |
0.51 |
Binding ≤ 10μM
|
|
DAPK3-2-E |
Death-associated Protein Kinase 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
410 |
0.53 |
Binding ≤ 10μM
|
|
E2AK2-1-E |
Interferon-induced, Double-stranded RNA-activated Protein Kinase (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
8000 |
0.42 |
Binding ≤ 10μM |
|
EPHA3-2-E |
Ephrin Type-A Receptor 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4000 |
0.44 |
Binding ≤ 10μM
|
|
GAK-2-E |
Serine/threonine-protein Kinase GAK (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
38 |
0.61 |
Binding ≤ 10μM
|
|
JAK3-2-E |
Tyrosine-protein Kinase JAK3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
957 |
0.50 |
Binding ≤ 10μM
|
|
KC1E-2-E |
Casein Kinase I Epsilon (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3100 |
0.45 |
Binding ≤ 10μM
|
|
KC1G1-2-E |
Casein Kinase I Gamma 1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
3700 |
0.45 |
Binding ≤ 10μM
|
|
KCC1A-2-E |
CaM Kinase I Alpha (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4100 |
0.44 |
Binding ≤ 10μM
|
|
KCC1D-2-E |
CaM Kinase I Delta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1400 |
0.48 |
Binding ≤ 10μM
|
|
KCC1G-2-E |
CaM Kinase I Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
4200 |
0.44 |
Binding ≤ 10μM
|
|
KKCC2-2-E |
CaM-kinase Kinase Beta (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
7500 |
0.42 |
Binding ≤ 10μM
|
|
KS6A2-2-E |
Ribosomal Protein S6 Kinase Alpha 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2200 |
0.47 |
Binding ≤ 10μM
|
|
KS6A5-1-E |
Ribosomal Protein S6 Kinase Alpha 5 (cluster #1 Of 3), Eukaryotic |
Eukaryotes |
1700 |
0.48 |
Binding ≤ 10μM
|
|
MARK2-2-E |
MAP/microtubule Affinity-regulating Kinase 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1800 |
0.47 |
Binding ≤ 10μM
|
|
MK08-5-E |
Mitogen-activated Protein Kinase 8 (cluster #5 Of 5), Eukaryotic |
Eukaryotes |
40 |
0.61 |
Binding ≤ 10μM
|
|
MK09-3-E |
C-Jun N-terminal Kinase 2 (cluster #3 Of 3), Eukaryotic |
Eukaryotes |
84 |
0.58 |
Binding ≤ 10μM
|
|
MK10-2-E |
C-Jun N-terminal Kinase 3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
90 |
0.58 |
Binding ≤ 10μM
|
|
MK12-2-E |
MAP Kinase P38 Gamma (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2400 |
0.46 |
Binding ≤ 10μM
|
|
MKNK2-2-E |
MAP Kinase Signal-integrating Kinase 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
1000 |
0.49 |
Binding ≤ 10μM
|
|
PHKG2-2-E |
Phosphorylase Kinase Gamma Subunit 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
480 |
0.52 |
Binding ≤ 10μM
|
|
PIM2-2-E |
Serine/threonine-protein Kinase PIM2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
460 |
0.52 |
Binding ≤ 10μM
|
|
PLK4-1-E |
Serine/threonine-protein Kinase PLK4 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
770 |
0.50 |
Binding ≤ 10μM
|
|
SLK-2-E |
Serine/threonine-protein Kinase 2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2300 |
0.46 |
Binding ≤ 10μM
|
|
ST17A-2-E |
Serine/threonine-protein Kinase 17A (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2100 |
0.47 |
Binding ≤ 10μM
|
|
ST38L-2-E |
Serine/threonine-protein Kinase 38-like (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2300 |
0.46 |
Binding ≤ 10μM
|
|
STK16-1-E |
Serine/threonine-protein Kinase 16 (cluster #1 Of 1), Eukaryotic |
Eukaryotes |
510 |
0.52 |
Binding ≤ 10μM
|
|
STK3-2-E |
Serine/threonine-protein Kinase MST2 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
220 |
0.55 |
Binding ≤ 10μM
|
|
STK4-2-E |
Serine/threonine-protein Kinase MST1 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
2200 |
0.47 |
Binding ≤ 10μM
|
|
TNIK-2-E |
TRAF2- And NCK-interacting Kinase (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
480 |
0.52 |
Binding ≤ 10μM
|
|
TTK-2-E |
Dual Specificity Protein Kinase TTK (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
98 |
0.58 |
Binding ≤ 10μM
|
|
ULK3-2-E |
Serine/threonine-protein Kinase ULK3 (cluster #2 Of 2), Eukaryotic |
Eukaryotes |
680 |
0.51 |
Binding ≤ 10μM
|
|
Z50425-3-O |
Plasmodium Falciparum (cluster #3 Of 22), Other |
Other |
7943 |
0.42 |
Functional ≤ 10μM
|
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
AAK1_HUMAN |
Q2M2I8
|
Adaptor-associated Kinase, Human |
35 |
0.61 |
Binding ≤ 1μM
|
|
BMP2K_HUMAN |
Q9NSY1
|
BMP-2-inducible Protein Kinase, Human |
25 |
0.63 |
Binding ≤ 1μM
|
|
MK08_HUMAN |
P45983
|
C-Jun N-terminal Kinase 1, Human |
100 |
0.58 |
Binding ≤ 1μM
|
|
MK09_HUMAN |
P45984
|
C-Jun N-terminal Kinase 2, Human |
40 |
0.61 |
Binding ≤ 1μM
|
|
MK10_HUMAN |
P53779
|
C-Jun N-terminal Kinase 3, Human |
22 |
0.63 |
Binding ≤ 1μM
|
|
DAPK2_HUMAN |
Q9UIK4
|
Death-associated Protein Kinase 2, Human |
610 |
0.51 |
Binding ≤ 1μM
|
|
DAPK3_HUMAN |
O43293
|
Death-associated Protein Kinase 3, Human |
410 |
0.53 |
Binding ≤ 1μM
|
|
CLK2_HUMAN |
P49760
|
Dual Specificity Protein Kinase CLK2, Human |
300 |
0.54 |
Binding ≤ 1μM
|
|
CLK4_HUMAN |
Q9HAZ1
|
Dual Specificity Protein Kinase CLK4, Human |
260 |
0.54 |
Binding ≤ 1μM
|
|
TTK_HUMAN |
P33981
|
Dual Specificity Protein Kinase TTK, Human |
98 |
0.58 |
Binding ≤ 1μM
|
|
MKNK2_HUMAN |
Q9HBH9
|
MAP Kinase Signal-integrating Kinase 2, Human |
1000 |
0.49 |
Binding ≤ 1μM
|
|
PHKG2_HUMAN |
P15735
|
Phosphorylase Kinase Gamma Subunit 2, Human |
480 |
0.52 |
Binding ≤ 1μM
|
|
STK16_HUMAN |
O75716
|
Serine/threonine-protein Kinase 16, Human |
510 |
0.52 |
Binding ≤ 1μM
|
|
AURKA_HUMAN |
O14965
|
Serine/threonine-protein Kinase Aurora-A, Human |
980 |
0.49 |
Binding ≤ 1μM
|
|
GAK_HUMAN |
O14976
|
Serine/threonine-protein Kinase GAK, Human |
38 |
0.61 |
Binding ≤ 1μM
|
|
STK3_MOUSE |
Q9JI10
|
Serine/threonine-protein Kinase MST2, Mouse |
220 |
0.55 |
Binding ≤ 1μM
|
|
CDK16_HUMAN |
Q00536
|
Serine/threonine-protein Kinase PCTAIRE-1, Human |
180 |
0.56 |
Binding ≤ 1μM
|
|
PIM2_HUMAN |
Q9P1W9
|
Serine/threonine-protein Kinase PIM2, Human |
460 |
0.52 |
Binding ≤ 1μM
|
|
PLK4_HUMAN |
O00444
|
Serine/threonine-protein Kinase PLK4, Human |
770 |
0.50 |
Binding ≤ 1μM
|
|
TNIK_HUMAN |
Q9UKE5
|
TRAF2- And NCK-interacting Kinase, Human |
480 |
0.52 |
Binding ≤ 1μM
|
|
JAK3_HUMAN |
P52333
|
Tyrosine-protein Kinase JAK3, Human |
957 |
0.50 |
Binding ≤ 1μM
|
|
ULK3_MOUSE |
Q3U3Q1
|
ULK3 Kinase, Mouse |
680 |
0.51 |
Binding ≤ 1μM
|
|
AAK1_HUMAN |
Q2M2I8
|
Adaptor-associated Kinase, Human |
35 |
0.61 |
Binding ≤ 10μM
|
|
BMP2K_HUMAN |
Q9NSY1
|
BMP-2-inducible Protein Kinase, Human |
25 |
0.63 |
Binding ≤ 10μM
|
|
MK08_HUMAN |
P45983
|
C-Jun N-terminal Kinase 1, Human |
100 |
0.58 |
Binding ≤ 10μM
|
|
MK09_HUMAN |
P45984
|
C-Jun N-terminal Kinase 2, Human |
40 |
0.61 |
Binding ≤ 10μM
|
|
MK10_HUMAN |
P53779
|
C-Jun N-terminal Kinase 3, Human |
22 |
0.63 |
Binding ≤ 10μM
|
|
KCC1A_HUMAN |
Q14012
|
CaM Kinase I Alpha, Human |
4100 |
0.44 |
Binding ≤ 10μM
|
|
KCC1D_HUMAN |
Q8IU85
|
CaM Kinase I Delta, Human |
1400 |
0.48 |
Binding ≤ 10μM
|
|
KCC1G_HUMAN |
Q96NX5
|
CaM Kinase I Gamma, Human |
4200 |
0.44 |
Binding ≤ 10μM
|
|
KKCC2_HUMAN |
Q96RR4
|
CaM-kinase Kinase Beta, Human |
7500 |
0.42 |
Binding ≤ 10μM
|
|
KC1E_HUMAN |
P49674
|
Casein Kinase I Epsilon, Human |
3100 |
0.45 |
Binding ≤ 10μM
|
|
KC1G1_HUMAN |
Q9HCP0
|
Casein Kinase I Gamma 1, Human |
3700 |
0.45 |
Binding ≤ 10μM
|
|
CDK2_HUMAN |
P24941
|
Cyclin-dependent Kinase 2, Human |
3200 |
0.45 |
Binding ≤ 10μM
|
|
DAPK2_HUMAN |
Q9UIK4
|
Death-associated Protein Kinase 2, Human |
610 |
0.51 |
Binding ≤ 10μM
|
|
DAPK3_HUMAN |
O43293
|
Death-associated Protein Kinase 3, Human |
410 |
0.53 |
Binding ≤ 10μM
|
|
CLK2_HUMAN |
P49760
|
Dual Specificity Protein Kinase CLK2, Human |
300 |
0.54 |
Binding ≤ 10μM
|
|
CLK4_HUMAN |
Q9HAZ1
|
Dual Specificity Protein Kinase CLK4, Human |
260 |
0.54 |
Binding ≤ 10μM
|
|
TTK_HUMAN |
P33981
|
Dual Specificity Protein Kinase TTK, Human |
98 |
0.58 |
Binding ≤ 10μM
|
|
EPHA3_HUMAN |
P29320
|
Ephrin Type-A Receptor 3, Human |
4000 |
0.44 |
Binding ≤ 10μM
|
|
E2AK2_HUMAN |
P19525
|
Interferon-induced, Double-stranded RNA-activated Protein Kinase, Human |
8000 |
0.42 |
Binding ≤ 10μM |
|
MK12_HUMAN |
P53778
|
MAP Kinase P38 Gamma, Human |
2400 |
0.46 |
Binding ≤ 10μM
|
|
MKNK2_HUMAN |
Q9HBH9
|
MAP Kinase Signal-integrating Kinase 2, Human |
1000 |
0.49 |
Binding ≤ 10μM
|
|
MARK2_HUMAN |
Q7KZI7
|
MAP/microtubule Affinity-regulating Kinase 2, Human |
1800 |
0.47 |
Binding ≤ 10μM
|
|
PHKG2_HUMAN |
P15735
|
Phosphorylase Kinase Gamma Subunit 2, Human |
480 |
0.52 |
Binding ≤ 10μM
|
|
KS6A2_HUMAN |
Q15349
|
Ribosomal Protein S6 Kinase Alpha 2, Human |
2200 |
0.47 |
Binding ≤ 10μM
|
|
KS6A5_HUMAN |
O75582
|
Ribosomal Protein S6 Kinase Alpha 5, Human |
1700 |
0.48 |
Binding ≤ 10μM
|
|
STK16_HUMAN |
O75716
|
Serine/threonine-protein Kinase 16, Human |
510 |
0.52 |
Binding ≤ 10μM
|
|
ST17A_HUMAN |
Q9UEE5
|
Serine/threonine-protein Kinase 17A, Human |
2100 |
0.47 |
Binding ≤ 10μM
|
|
SLK_HUMAN |
Q9H2G2
|
Serine/threonine-protein Kinase 2, Human |
2300 |
0.46 |
Binding ≤ 10μM
|
|
ST38L_HUMAN |
Q9Y2H1
|
Serine/threonine-protein Kinase 38-like, Human |
2300 |
0.46 |
Binding ≤ 10μM
|
|
AURKA_HUMAN |
O14965
|
Serine/threonine-protein Kinase Aurora-A, Human |
980 |
0.49 |
Binding ≤ 10μM
|
|
AURKC_HUMAN |
Q9UQB9
|
Serine/threonine-protein Kinase Aurora-C, Human |
5300 |
0.43 |
Binding ≤ 10μM
|
|
GAK_HUMAN |
O14976
|
Serine/threonine-protein Kinase GAK, Human |
38 |
0.61 |
Binding ≤ 10μM
|
|
STK4_HUMAN |
Q13043
|
Serine/threonine-protein Kinase MST1, Human |
2200 |
0.47 |
Binding ≤ 10μM
|
|
STK3_MOUSE |
Q9JI10
|
Serine/threonine-protein Kinase MST2, Mouse |
220 |
0.55 |
Binding ≤ 10μM
|
|
CDK16_HUMAN |
Q00536
|
Serine/threonine-protein Kinase PCTAIRE-1, Human |
180 |
0.56 |
Binding ≤ 10μM
|
|
PIM2_HUMAN |
Q9P1W9
|
Serine/threonine-protein Kinase PIM2, Human |
460 |
0.52 |
Binding ≤ 10μM
|
|
PLK4_HUMAN |
O00444
|
Serine/threonine-protein Kinase PLK4, Human |
770 |
0.50 |
Binding ≤ 10μM
|
|
TNIK_HUMAN |
Q9UKE5
|
TRAF2- And NCK-interacting Kinase, Human |
480 |
0.52 |
Binding ≤ 10μM
|
|
JAK3_HUMAN |
P52333
|
Tyrosine-protein Kinase JAK3, Human |
957 |
0.50 |
Binding ≤ 10μM
|
|
ULK3_MOUSE |
Q3U3Q1
|
ULK3 Kinase, Mouse |
680 |
0.51 |
Binding ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
7943.28235 |
0.42 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.48 |
6.16 |
-10.65 |
1 |
3 |
0 |
46 |
220.231 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.49 |
4.15 |
-24.93 |
1 |
3 |
0 |
46 |
220.231 |
0 |
↓
|
|
Mid
Mid (pH 6-8)
|
3.48 |
6.17 |
-10.96 |
1 |
3 |
0 |
46 |
220.231 |
0 |
↓
|
|
|
|
Analogs
-
89382
-
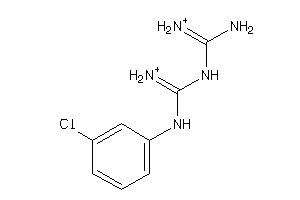
Draw
Identity
99%
90%
80%
70%
ChEMBL Target Annotations
| Uniprot |
Swissprot |
Affinity (nM) |
LE (kcal/mol/atom) |
Type |
|
5HT3A_HUMAN |
P46098
|
Serotonin 3a (5-HT3a) Receptor, Human |
17 |
0.78 |
Binding ≤ 1μM
|
|
5HT3B_HUMAN |
O95264
|
Serotonin 3b (5-HT3b) Receptor, Human |
17 |
0.78 |
Binding ≤ 1μM
|
|
5HT3C_HUMAN |
Q8WXA8
|
Serotonin 3c (5-HT3c) Receptor, Human |
17 |
0.78 |
Binding ≤ 1μM
|
|
5HT3D_HUMAN |
Q70Z44
|
Serotonin 3d (5-HT3d) Receptor, Human |
17 |
0.78 |
Binding ≤ 1μM
|
|
5HT3E_HUMAN |
A5X5Y0
|
Serotonin 3e (5-HT3e) Receptor, Human |
17 |
0.78 |
Binding ≤ 1μM
|
|
5HT3A_HUMAN |
P46098
|
Serotonin 3a (5-HT3a) Receptor, Human |
17 |
0.78 |
Binding ≤ 10μM
|
|
5HT3B_HUMAN |
O95264
|
Serotonin 3b (5-HT3b) Receptor, Human |
17 |
0.78 |
Binding ≤ 10μM
|
|
5HT3C_HUMAN |
Q8WXA8
|
Serotonin 3c (5-HT3c) Receptor, Human |
17 |
0.78 |
Binding ≤ 10μM
|
|
5HT3D_HUMAN |
Q70Z44
|
Serotonin 3d (5-HT3d) Receptor, Human |
17 |
0.78 |
Binding ≤ 10μM
|
|
5HT3E_HUMAN |
A5X5Y0
|
Serotonin 3e (5-HT3e) Receptor, Human |
17 |
0.78 |
Binding ≤ 10μM
|
|
Z50425 |
Z50425
|
Plasmodium Falciparum |
5011.87234 |
0.53 |
Functional ≤ 10μM
|
|
5HT3A_MOUSE |
P23979
|
Serotonin 3a (5-HT3a) Receptor, Mouse |
800 |
0.61 |
Functional ≤ 10μM
|
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
0.85 |
5.79 |
-36.36 |
7 |
5 |
1 |
102 |
212.664 |
3 |
↓
|
|
Ref
Reference (pH 7)
|
0.98 |
5.93 |
-28.51 |
7 |
5 |
1 |
100 |
212.664 |
4 |
↓
|
|
Mid
Mid (pH 6-8)
|
0.98 |
5.94 |
-5.07 |
6 |
5 |
0 |
98 |
211.656 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
3.06 |
6.04 |
-11.94 |
2 |
6 |
0 |
87 |
295.298 |
3 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Ref
Reference (pH 7)
|
2.92 |
5.45 |
-8.46 |
2 |
5 |
0 |
70 |
265.338 |
4 |
↓
|
|
Ref
Reference (pH 7)
|
2.92 |
5.45 |
-9.77 |
2 |
5 |
0 |
70 |
265.338 |
4 |
↓
|
|
|
|
Analogs
Draw
Identity
99%
90%
80%
70%
Physical Representations
|
Type
pH range
|
xlogP
|
Des A‑Pol
Apolar desolvation
(kcal/mol)
|
Des Pol
Polar desolvation
(kcal/mol)
|
H Don
H-bond donors
|
H Acc
H-bond acceptors
|
Chg
Net charge
|
tPSA
(Ų)
|
MWT
Molecular weight
(g/mol)
|
RB
Rotatable bonds
|
DL |
|
Mid
Mid (pH 6-8)
|
1.65 |
3.61 |
-56.38 |
2 |
8 |
-1 |
132 |
354.389 |
6 |
↓
|
|
Lo
Low (pH 4.5-6)
|
1.65 |
1.63 |
-20.55 |
3 |
8 |
0 |
129 |
355.397 |
6 |
↓
|
|